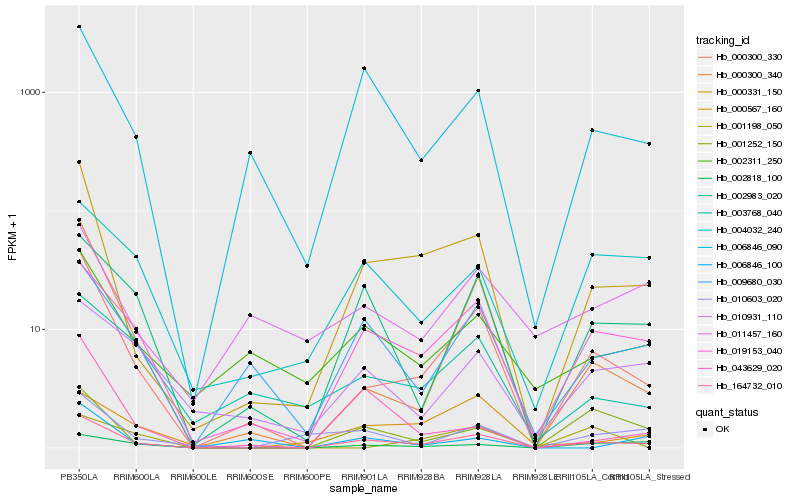
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_330 |
0.0 |
- |
- |
- |
| 2 |
Hb_000300_340 |
0.136935957 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_164732_010 |
0.1529180436 |
- |
- |
- |
| 4 |
Hb_000567_160 |
0.1879314657 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 5 |
Hb_019153_040 |
0.2167629749 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_009680_030 |
0.2491605225 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 7 |
Hb_002983_020 |
0.2643661182 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Populus euphratica] |
| 8 |
Hb_003768_040 |
0.2665765152 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_006846_090 |
0.2724874026 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 10 |
Hb_002311_250 |
0.2767871778 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 11 |
Hb_001252_150 |
0.2922155733 |
- |
- |
- |
| 12 |
Hb_010931_110 |
0.2928485289 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_002818_100 |
0.2990398186 |
- |
- |
hypothetical protein JCGZ_14122 [Jatropha curcas] |
| 14 |
Hb_010603_020 |
0.301764914 |
- |
- |
- |
| 15 |
Hb_001198_050 |
0.3090325266 |
- |
- |
hypothetical protein POPTR_0004s22990g [Populus trichocarpa] |
| 16 |
Hb_004032_240 |
0.311838464 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 17 |
Hb_000331_150 |
0.3156871548 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_043629_020 |
0.3172515357 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 19 |
Hb_011457_160 |
0.3185420984 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 20 |
Hb_006846_100 |
0.3190864878 |
- |
- |
hypothetical protein POPTR_0012s11270g [Populus trichocarpa] |