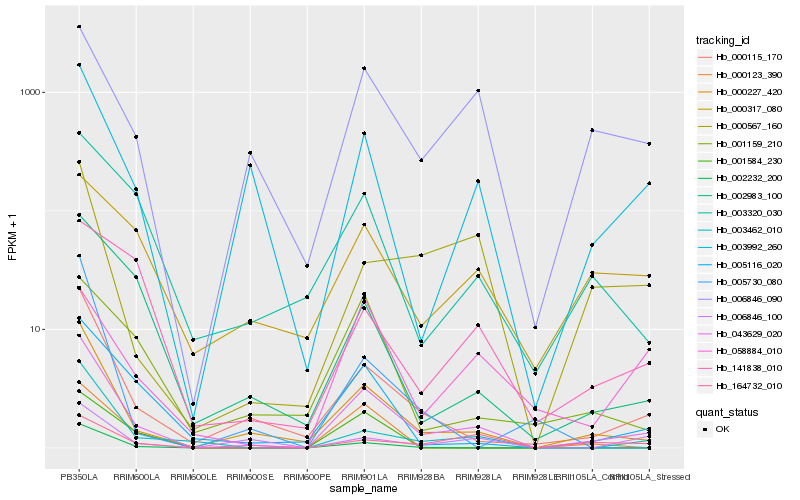
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043629_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 2 |
Hb_000227_420 |
0.1327480114 |
- |
- |
PREDICTED: thermospermine synthase ACAULIS5-like [Jatropha curcas] |
| 3 |
Hb_000115_170 |
0.1727838352 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 4 |
Hb_003462_010 |
0.205687058 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_003992_260 |
0.2176350726 |
- |
- |
PREDICTED: 26.5 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 6 |
Hb_005116_020 |
0.2179906369 |
- |
- |
PREDICTED: DNA repair helicase UVH6 isoform X2 [Sesamum indicum] |
| 7 |
Hb_002232_200 |
0.2268929362 |
- |
- |
PREDICTED: sugar carrier protein C-like [Jatropha curcas] |
| 8 |
Hb_006846_090 |
0.2449362488 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 9 |
Hb_002983_100 |
0.2454541129 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF084 [Jatropha curcas] |
| 10 |
Hb_003320_030 |
0.2462604154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000123_390 |
0.2597258917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_164732_010 |
0.261816121 |
- |
- |
- |
| 13 |
Hb_006846_100 |
0.2643069137 |
- |
- |
hypothetical protein POPTR_0012s11270g [Populus trichocarpa] |
| 14 |
Hb_000567_160 |
0.2681260922 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 15 |
Hb_001159_210 |
0.2759861349 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 16 |
Hb_001584_230 |
0.2765159365 |
- |
- |
invertase/pectin methylesterase inhibitor family protein [Populus trichocarpa] |
| 17 |
Hb_058884_010 |
0.2777169021 |
- |
- |
PREDICTED: barwin-like [Jatropha curcas] |
| 18 |
Hb_005730_080 |
0.2802996083 |
- |
- |
PREDICTED: 17.6 kDa class I heat shock protein-like [Solanum lycopersicum] |
| 19 |
Hb_141838_010 |
0.2819649681 |
- |
- |
- |
| 20 |
Hb_000317_080 |
0.2820749638 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |