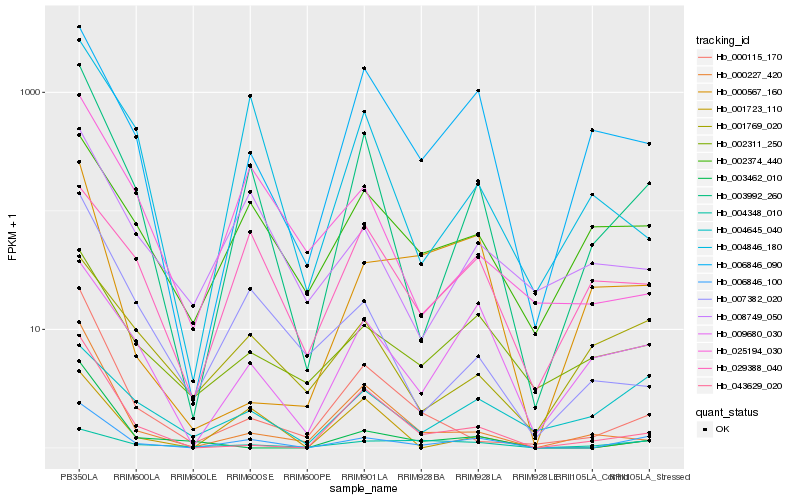
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s11270g [Populus trichocarpa] |
| 2 |
Hb_003992_260 |
0.17269591 |
- |
- |
PREDICTED: 26.5 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 3 |
Hb_009680_030 |
0.2519009826 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 4 |
Hb_043629_020 |
0.2643069137 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 5 |
Hb_006846_090 |
0.2658861912 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 6 |
Hb_002311_250 |
0.2678000276 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 7 |
Hb_000115_170 |
0.2774643179 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 8 |
Hb_004645_040 |
0.2777226237 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_002374_440 |
0.2805825738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008749_050 |
0.2823263539 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 11 |
Hb_001723_110 |
0.2832485439 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 12 |
Hb_007382_020 |
0.2872700978 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 13 |
Hb_003462_010 |
0.2886925562 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_000227_420 |
0.289708228 |
- |
- |
PREDICTED: thermospermine synthase ACAULIS5-like [Jatropha curcas] |
| 15 |
Hb_004348_010 |
0.289710924 |
- |
- |
hypothetical protein POPTR_0077s002302g, partial [Populus trichocarpa] |
| 16 |
Hb_001769_020 |
0.2900248383 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_029388_040 |
0.2968709287 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 18 |
Hb_025194_030 |
0.2970447909 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 19 |
Hb_000567_160 |
0.298197882 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 20 |
Hb_004846_180 |
0.2997268786 |
- |
- |
heat shock protein, putative [Ricinus communis] |