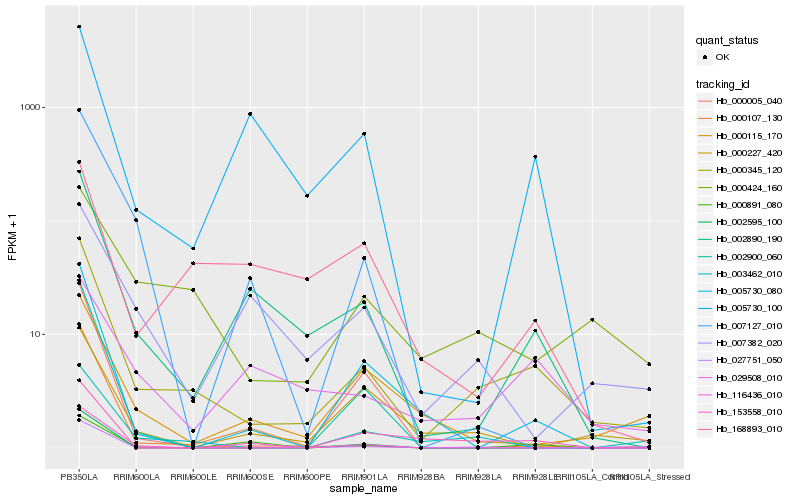
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_120 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |
| 2 |
Hb_002890_190 |
0.2211101403 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 3 |
Hb_000424_160 |
0.2285272964 |
- |
- |
hypothetical protein POPTR_0006s23570g [Populus trichocarpa] |
| 4 |
Hb_003462_010 |
0.2300370086 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_005730_080 |
0.2495895383 |
- |
- |
PREDICTED: 17.6 kDa class I heat shock protein-like [Solanum lycopersicum] |
| 6 |
Hb_027751_050 |
0.256504436 |
- |
- |
hypothetical protein JCGZ_05887 [Jatropha curcas] |
| 7 |
Hb_153558_010 |
0.2611634599 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 8 |
Hb_029508_010 |
0.2645925723 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002900_060 |
0.2703437224 |
- |
- |
CI small heat shock protein 2 [Prunus salicina] |
| 10 |
Hb_000227_420 |
0.2707863373 |
- |
- |
PREDICTED: thermospermine synthase ACAULIS5-like [Jatropha curcas] |
| 11 |
Hb_000115_170 |
0.2709550258 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 12 |
Hb_000107_130 |
0.2737633659 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 13 |
Hb_005730_100 |
0.2750586739 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 14 |
Hb_000005_040 |
0.2776317189 |
- |
- |
PREDICTED: histone H2B.2 [Jatropha curcas] |
| 15 |
Hb_007127_010 |
0.2804428115 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 16 |
Hb_000891_080 |
0.280841008 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 17 |
Hb_007382_020 |
0.2833472757 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 18 |
Hb_116436_010 |
0.2870124295 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 19 |
Hb_002595_100 |
0.2900614131 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 20 |
Hb_168893_010 |
0.2930726421 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |