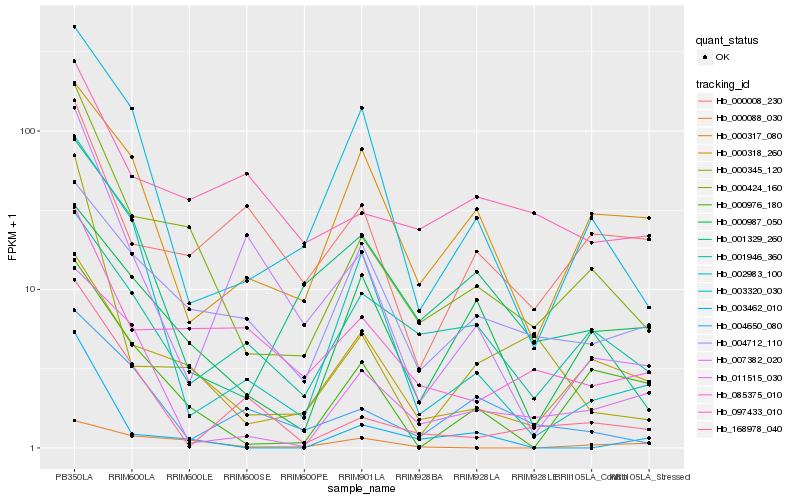
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_160 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s23570g [Populus trichocarpa] |
| 2 |
Hb_000318_260 |
0.1769820731 |
- |
- |
PREDICTED: cell division cycle-associated 7-like protein [Jatropha curcas] |
| 3 |
Hb_085375_010 |
0.2016415134 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000976_180 |
0.2217215211 |
- |
- |
PREDICTED: uncharacterized protein LOC102592049 [Solanum tuberosum] |
| 5 |
Hb_000345_120 |
0.2285272964 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |
| 6 |
Hb_003320_030 |
0.2355872133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004712_110 |
0.2418562491 |
- |
- |
30S ribosomal protein S10, putative [Ricinus communis] |
| 8 |
Hb_097433_010 |
0.2460002683 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 9 |
Hb_001329_260 |
0.2470005371 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000987_050 |
0.2483797659 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003462_010 |
0.2503536667 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_011515_030 |
0.2536088371 |
- |
- |
- |
| 13 |
Hb_001946_360 |
0.255771423 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 14 |
Hb_002983_100 |
0.2566294703 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF084 [Jatropha curcas] |
| 15 |
Hb_168978_040 |
0.2602143933 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 16 |
Hb_000008_230 |
0.2615882116 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 17 |
Hb_000088_030 |
0.2636510029 |
- |
- |
hypothetical protein JCGZ_10947 [Jatropha curcas] |
| 18 |
Hb_004650_080 |
0.2675897619 |
- |
- |
- |
| 19 |
Hb_007382_020 |
0.2695628632 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 20 |
Hb_000317_080 |
0.2717545218 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |