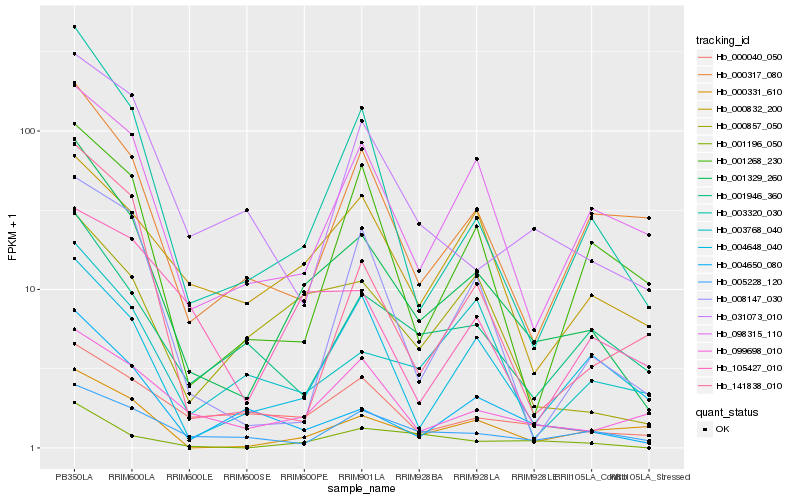
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003320_030 |
0.1586456166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004650_080 |
0.1897276727 |
- |
- |
- |
| 4 |
Hb_001196_050 |
0.1974410026 |
- |
- |
- |
| 5 |
Hb_000857_050 |
0.2067162323 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 6 |
Hb_098315_110 |
0.2072566992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000331_610 |
0.2097763224 |
- |
- |
hypothetical protein B456_003G108600 [Gossypium raimondii] |
| 8 |
Hb_000317_080 |
0.2122996491 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 9 |
Hb_001946_360 |
0.2145633486 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 10 |
Hb_004648_040 |
0.2145927423 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 11 |
Hb_141838_010 |
0.2151920504 |
- |
- |
- |
| 12 |
Hb_008147_030 |
0.219306572 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 13 |
Hb_099698_010 |
0.2230210855 |
- |
- |
- |
| 14 |
Hb_031073_010 |
0.2241203955 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 15 |
Hb_000832_200 |
0.2261786542 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000040_050 |
0.2262733164 |
- |
- |
PREDICTED: uncharacterized protein LOC102617857 isoform X1 [Citrus sinensis] |
| 17 |
Hb_001268_230 |
0.2297212735 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003768_040 |
0.2307627842 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_105427_010 |
0.2333271474 |
- |
- |
- |
| 20 |
Hb_005228_120 |
0.2337350508 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |