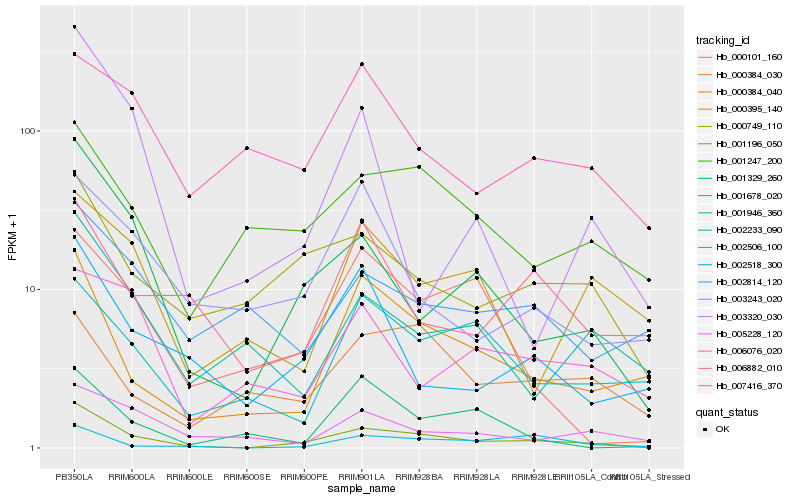
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001196_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_001329_260 |
0.1974410026 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000384_040 |
0.2317617104 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 4 |
Hb_000749_110 |
0.2373669872 |
- |
- |
PREDICTED: choline transporter-like protein 2 [Jatropha curcas] |
| 5 |
Hb_002518_300 |
0.2528587598 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001247_200 |
0.2570949539 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 7 |
Hb_002506_100 |
0.2589823658 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001946_360 |
0.2602479634 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 9 |
Hb_005228_120 |
0.262369628 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 10 |
Hb_000101_160 |
0.2639793979 |
- |
- |
PREDICTED: uncharacterized protein LOC105133607 isoform X1 [Populus euphratica] |
| 11 |
Hb_003243_020 |
0.2756220824 |
- |
- |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 12 |
Hb_007416_370 |
0.2761424611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002233_090 |
0.2763062708 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_002814_120 |
0.2790098779 |
- |
- |
PREDICTED: uncharacterized protein LOC105633463 [Jatropha curcas] |
| 15 |
Hb_006882_010 |
0.281858208 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Cucumis sativus] |
| 16 |
Hb_000384_030 |
0.2841814835 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_003320_030 |
0.2843985314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006076_020 |
0.2845707095 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 19 |
Hb_001678_020 |
0.2846371102 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 20 |
Hb_000395_140 |
0.2854875284 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |