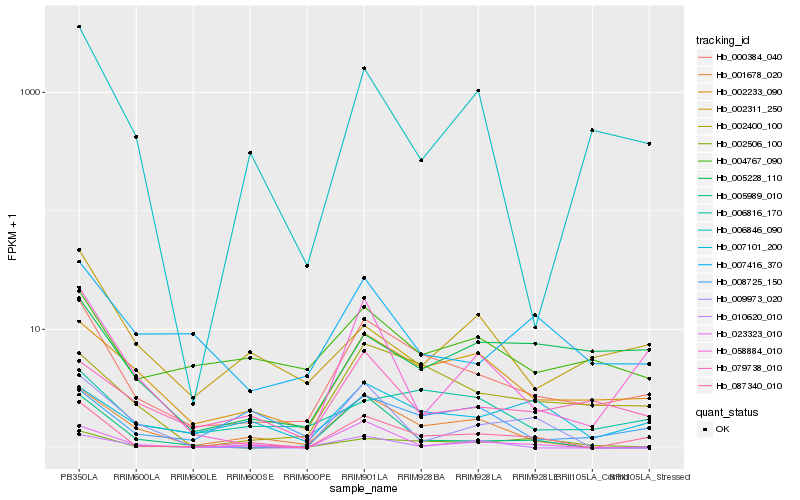
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087340_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000384_040 |
0.2007126766 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 3 |
Hb_058884_010 |
0.2468815512 |
- |
- |
PREDICTED: barwin-like [Jatropha curcas] |
| 4 |
Hb_001678_020 |
0.2537429556 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 5 |
Hb_007101_200 |
0.2723551459 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 6 |
Hb_023323_010 |
0.2729375004 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_002233_090 |
0.2735271585 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_006846_090 |
0.2975536347 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 9 |
Hb_002506_100 |
0.2997313798 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 10 |
Hb_008725_150 |
0.3000373976 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 11 |
Hb_002400_100 |
0.3021325748 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005228_110 |
0.3022691857 |
- |
- |
Cleavage and polyadenylation specificity factor subunit 3 [Fukomys damarensis] |
| 13 |
Hb_010620_010 |
0.3048471938 |
- |
- |
hypothetical protein JCGZ_22570 [Jatropha curcas] |
| 14 |
Hb_005989_010 |
0.307146 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 15 |
Hb_009973_020 |
0.3083906588 |
- |
- |
PREDICTED: uncharacterized protein LOC104889111 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_079738_010 |
0.3099750753 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 17 |
Hb_002311_250 |
0.3101582379 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 18 |
Hb_007416_370 |
0.3108296518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004767_090 |
0.3122831256 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 20 |
Hb_006816_170 |
0.313401776 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |