| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
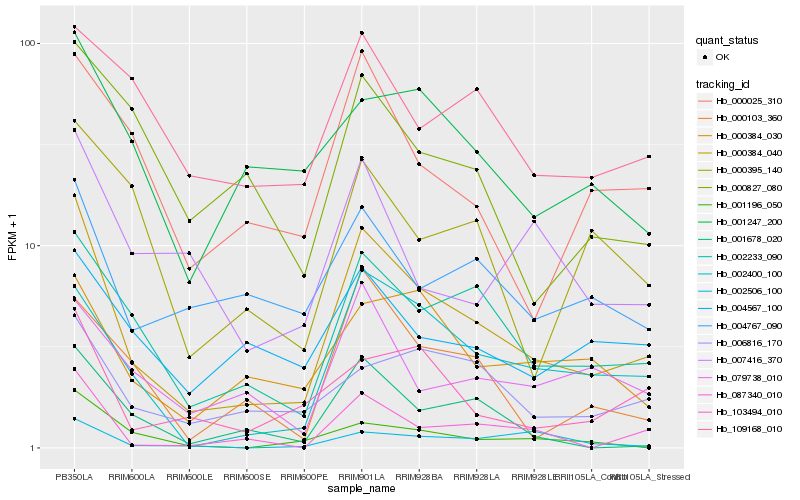
Hb_000384_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 2 |
Hb_002233_090 |
0.1623348865 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_002400_100 |
0.1770523648 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 4 |
Hb_087340_010 |
0.2007126766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000025_310 |
0.2060645987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_103494_010 |
0.2083911103 |
- |
- |
- |
| 7 |
Hb_001678_020 |
0.214701852 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 8 |
Hb_006816_170 |
0.2147954154 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 9 |
Hb_000103_360 |
0.216175192 |
- |
- |
- |
| 10 |
Hb_000384_030 |
0.2185506712 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_079738_010 |
0.2188321683 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 12 |
Hb_004767_090 |
0.2217208946 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001247_200 |
0.2236317658 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 14 |
Hb_004567_100 |
0.2256043474 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 15 |
Hb_000395_140 |
0.2290583243 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 16 |
Hb_007416_370 |
0.230243398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001196_050 |
0.2317617104 |
- |
- |
- |
| 18 |
Hb_000827_080 |
0.2318695473 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_109168_010 |
0.232338483 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_002506_100 |
0.2359066521 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |