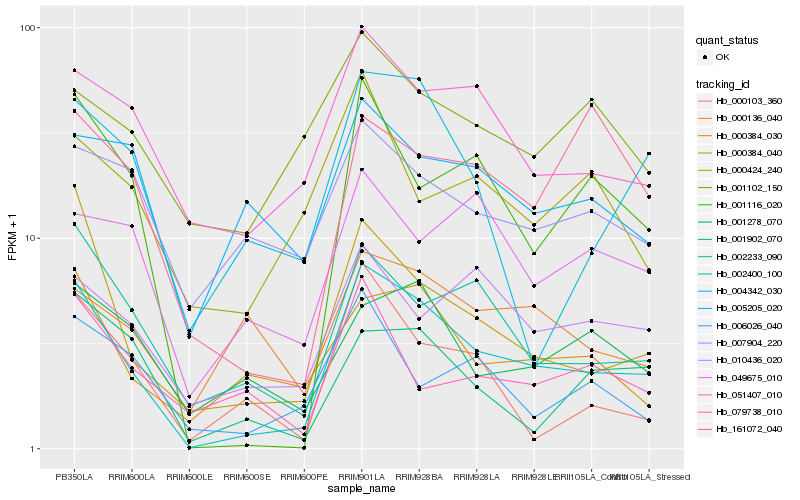
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_100 |
0.0 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000384_040 |
0.1770523648 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 3 |
Hb_001116_020 |
0.181244624 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2-like [Eucalyptus grandis] |
| 4 |
Hb_002233_090 |
0.1888836711 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_051407_010 |
0.2045825262 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_000384_030 |
0.2069055522 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_010436_020 |
0.2185613766 |
- |
- |
Dicer-like protein 4 [Morus notabilis] |
| 8 |
Hb_000103_360 |
0.2235667905 |
- |
- |
- |
| 9 |
Hb_004342_030 |
0.2249164599 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 10 |
Hb_001278_070 |
0.226428125 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 11 |
Hb_001902_070 |
0.2291036571 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_006026_040 |
0.2303967719 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 13 |
Hb_049675_010 |
0.230820582 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 14 |
Hb_007904_220 |
0.2332200996 |
- |
- |
PREDICTED: protein SDA1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_079738_010 |
0.2338872956 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 16 |
Hb_161072_040 |
0.2357719727 |
- |
- |
PREDICTED: replication factor C subunit 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000424_240 |
0.2362320267 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 18 |
Hb_005205_020 |
0.2362437612 |
- |
- |
PREDICTED: protein FAM135B-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000136_040 |
0.2381909166 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 20 |
Hb_001102_150 |
0.2417235695 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Jatropha curcas] |