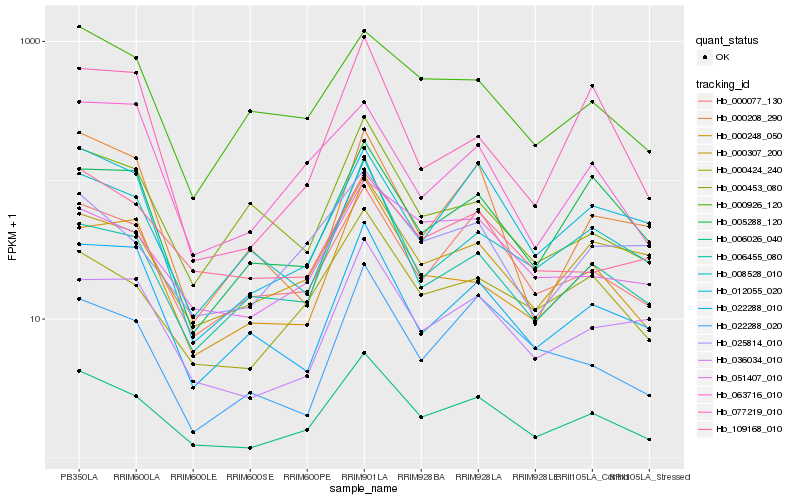
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006026_040 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 2 |
Hb_000424_240 |
0.1198903504 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 3 |
Hb_000077_130 |
0.1199718569 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 4 |
Hb_036034_010 |
0.1337435218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000248_050 |
0.1372536352 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 6 |
Hb_022288_010 |
0.139786232 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 7 |
Hb_000307_200 |
0.1423359936 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 8 |
Hb_012055_020 |
0.1431475845 |
- |
- |
PREDICTED: metal transporter Nramp3 [Jatropha curcas] |
| 9 |
Hb_006455_080 |
0.1460599353 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 10 |
Hb_051407_010 |
0.146890189 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_005288_120 |
0.1497041621 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 12 |
Hb_000926_120 |
0.1499604603 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 13 |
Hb_025814_010 |
0.1507058261 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 14 |
Hb_022288_020 |
0.1526719149 |
- |
- |
hypothetical protein POPTR_0006s08590g [Populus trichocarpa] |
| 15 |
Hb_109168_010 |
0.1543438433 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_063716_010 |
0.1548183944 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 17 |
Hb_008528_010 |
0.1562390297 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 18 |
Hb_000208_290 |
0.1563734302 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_077219_010 |
0.1569068274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000453_080 |
0.1570145055 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |