| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
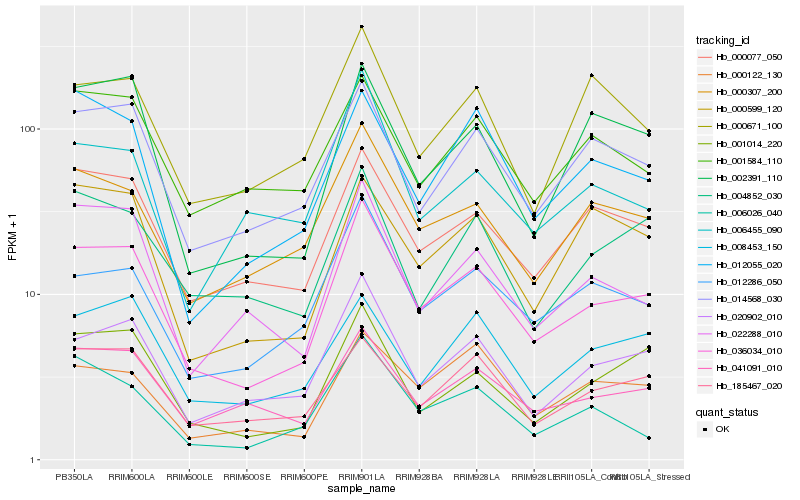
Hb_036034_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_020902_010 |
0.0931049007 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 3 |
Hb_000077_050 |
0.1071588267 |
- |
- |
PREDICTED: elongation factor G-1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000307_200 |
0.1075627341 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 5 |
Hb_012286_050 |
0.1105552305 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 6 |
Hb_014568_030 |
0.110891352 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 7 |
Hb_041091_010 |
0.1135312561 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001014_220 |
0.1139529738 |
- |
- |
PREDICTED: mediator-associated protein 2 [Jatropha curcas] |
| 9 |
Hb_022288_010 |
0.117046611 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 10 |
Hb_185467_020 |
0.1234406992 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 11 |
Hb_002391_110 |
0.1269725091 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_004852_030 |
0.1319335258 |
- |
- |
PREDICTED: uncharacterized protein LOC105639923 [Jatropha curcas] |
| 13 |
Hb_006026_040 |
0.1337435218 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 14 |
Hb_000122_130 |
0.1343527768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001584_110 |
0.1345534887 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000599_120 |
0.1352581027 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT34 [Jatropha curcas] |
| 17 |
Hb_006455_090 |
0.1355323556 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 18 |
Hb_008453_150 |
0.1358716299 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012055_020 |
0.1359483822 |
- |
- |
PREDICTED: metal transporter Nramp3 [Jatropha curcas] |
| 20 |
Hb_000671_100 |
0.136464056 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |