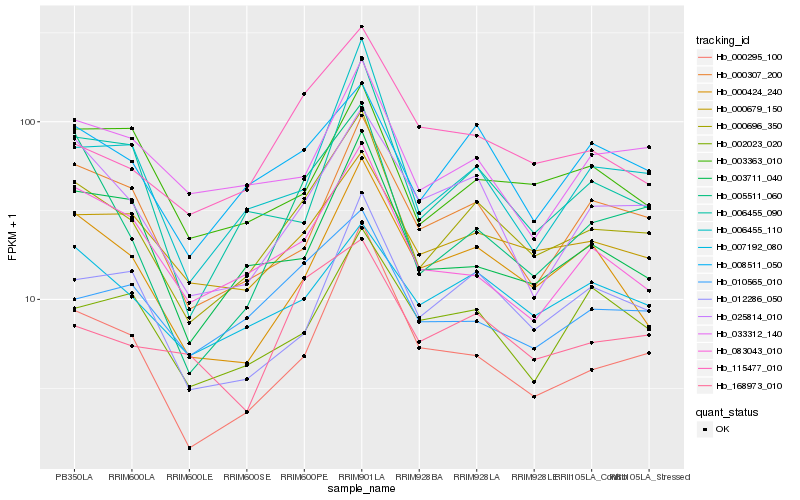
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_350 |
0.0 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_008511_050 |
0.1193869098 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 3 |
Hb_000679_150 |
0.1199367585 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_006455_110 |
0.1215177615 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_012286_050 |
0.1257689745 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 6 |
Hb_006455_090 |
0.126971822 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 7 |
Hb_025814_010 |
0.1275300028 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 8 |
Hb_000307_200 |
0.1278852149 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 9 |
Hb_000424_240 |
0.1299182142 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 10 |
Hb_010565_010 |
0.1322107441 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 11 |
Hb_033312_140 |
0.1345882998 |
- |
- |
RNA and export factor-binding family protein [Populus trichocarpa] |
| 12 |
Hb_003711_040 |
0.134871893 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_007192_080 |
0.1350346838 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005511_060 |
0.138309789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_083043_010 |
0.1388389159 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 16 |
Hb_115477_010 |
0.1413556455 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 17 |
Hb_000295_100 |
0.141520803 |
- |
- |
- |
| 18 |
Hb_003363_010 |
0.1432943169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_168973_010 |
0.1454172226 |
- |
- |
- |
| 20 |
Hb_002023_020 |
0.1482098942 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |