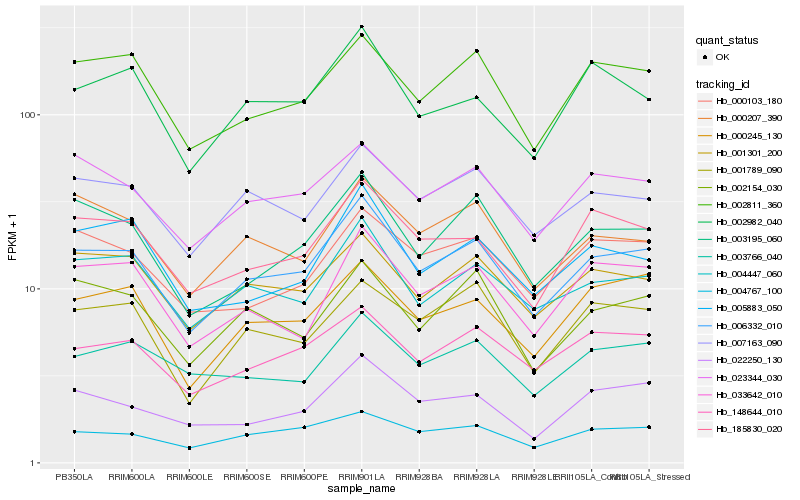
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006332_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 isoform X1 [Vitis vinifera] |
| 2 |
Hb_004447_060 |
0.0698825747 |
- |
- |
PREDICTED: uncharacterized protein LOC105636329 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007163_090 |
0.0743866248 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 4 |
Hb_148644_010 |
0.0761445912 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002811_360 |
0.0780279755 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 6 |
Hb_004767_100 |
0.0785645581 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_001301_200 |
0.0788201485 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 8 |
Hb_023344_030 |
0.078856465 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 9 |
Hb_003195_060 |
0.0788762576 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 10 |
Hb_185830_020 |
0.0794307006 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000245_130 |
0.0827253141 |
- |
- |
PREDICTED: nuclear pore complex protein NUP35 [Jatropha curcas] |
| 12 |
Hb_033642_010 |
0.0841725173 |
- |
- |
PREDICTED: protection of telomeres protein 1a-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000207_390 |
0.0852788233 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 14 |
Hb_002982_040 |
0.0862286236 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 15 |
Hb_022250_130 |
0.0868481621 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 16 |
Hb_005883_050 |
0.0878305243 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 17 |
Hb_002154_030 |
0.0884173598 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000103_180 |
0.0884299776 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 19 |
Hb_003766_040 |
0.0885783102 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001789_090 |
0.0893644182 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |