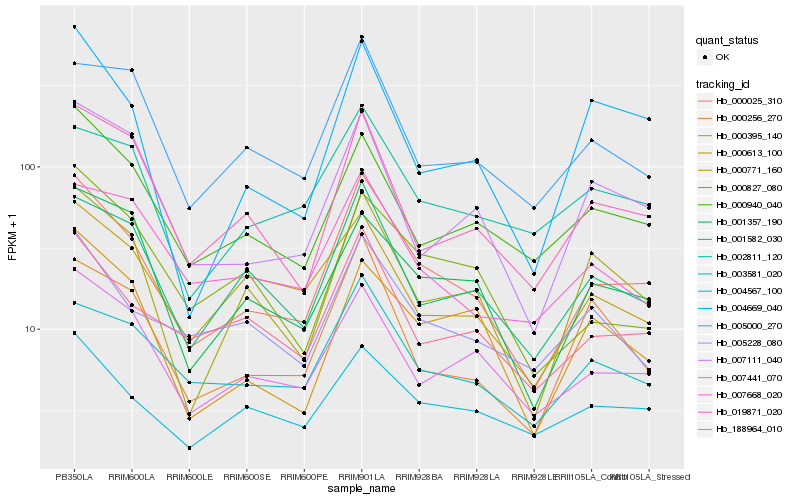
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_310 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_188964_010 |
0.1194322719 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 3 |
Hb_001582_030 |
0.1204373648 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 4 |
Hb_007441_070 |
0.1206234581 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 5 |
Hb_000771_160 |
0.1241071801 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 6 |
Hb_003581_020 |
0.1271511992 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 7 |
Hb_019871_020 |
0.1277190594 |
- |
- |
PREDICTED: uncharacterized protein LOC105642047 [Jatropha curcas] |
| 8 |
Hb_000940_040 |
0.127876755 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 9 |
Hb_004567_100 |
0.1344391694 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 10 |
Hb_001357_190 |
0.1346278816 |
- |
- |
ribonuclease III, putative [Ricinus communis] |
| 11 |
Hb_007111_040 |
0.1357061059 |
- |
- |
PREDICTED: uncharacterized protein LOC105628142 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005000_270 |
0.1390538746 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_004669_040 |
0.1392334396 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 14 |
Hb_005228_080 |
0.1399878028 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 15 |
Hb_000256_270 |
0.140097434 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 16 |
Hb_000395_140 |
0.1415452147 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 17 |
Hb_000827_080 |
0.1423621022 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000613_100 |
0.1425411592 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 19 |
Hb_002811_120 |
0.1435555115 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 20 |
Hb_007668_020 |
0.1453883357 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |