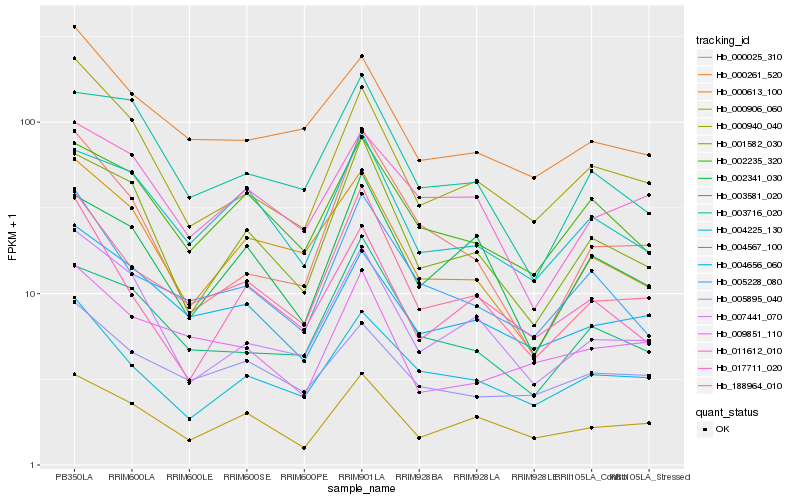
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_188964_010 |
0.0 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 2 |
Hb_005228_080 |
0.0918730997 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 3 |
Hb_003581_020 |
0.1098424295 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000906_060 |
0.1106206092 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 5 |
Hb_004567_100 |
0.1110053935 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 6 |
Hb_001582_030 |
0.1110880835 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 7 |
Hb_000613_100 |
0.1145246906 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 8 |
Hb_007441_070 |
0.1188260019 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 9 |
Hb_000261_520 |
0.1189056552 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 10 |
Hb_000025_310 |
0.1194322719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000940_040 |
0.1203254226 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 12 |
Hb_004225_130 |
0.120415476 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002341_030 |
0.1260691012 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 14 |
Hb_004656_060 |
0.1264441998 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 15 |
Hb_009851_110 |
0.1271906684 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 16 |
Hb_002235_320 |
0.1292598539 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011612_010 |
0.1309737511 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_005895_040 |
0.1333099485 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 19 |
Hb_017711_020 |
0.1336865473 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003716_020 |
0.1338106003 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |