| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
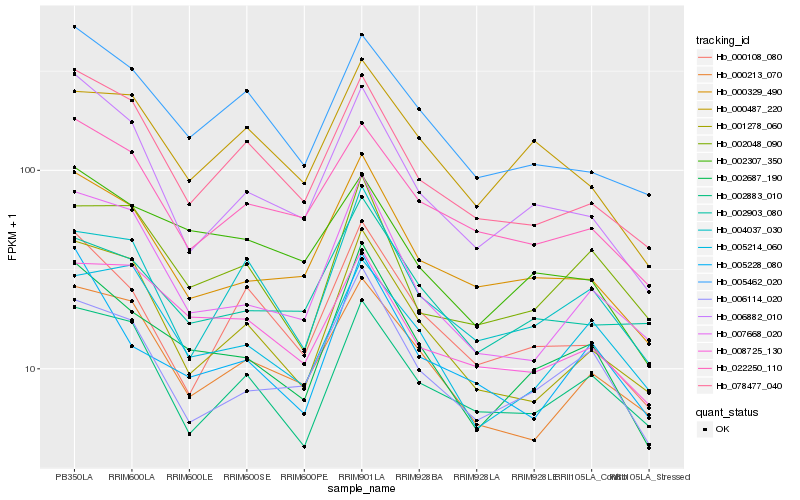
Hb_002687_190 |
0.0 |
- |
- |
prolyl oligopeptidase [Cicer arietinum] |
| 2 |
Hb_006114_020 |
0.1002663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000329_490 |
0.1009868386 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 4 |
Hb_007668_020 |
0.1108774013 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 5 |
Hb_005228_080 |
0.1131296237 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 6 |
Hb_002307_350 |
0.1136612287 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_006882_010 |
0.1172679094 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Cucumis sativus] |
| 8 |
Hb_001278_060 |
0.1178201629 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 9 |
Hb_005462_020 |
0.1206488789 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 10 |
Hb_000108_080 |
0.1244230699 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 11 |
Hb_002048_090 |
0.1245521835 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 12 |
Hb_008725_130 |
0.1251802408 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 13 |
Hb_000213_070 |
0.1254994299 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 14 |
Hb_078477_040 |
0.126210438 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 15 |
Hb_002903_080 |
0.1262788352 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 16 |
Hb_005214_060 |
0.1270316001 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_022250_110 |
0.1272195201 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_004037_030 |
0.1276449237 |
- |
- |
PREDICTED: DNA repair endonuclease UVH1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000487_220 |
0.1281765341 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 20 |
Hb_002883_010 |
0.1294049383 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |