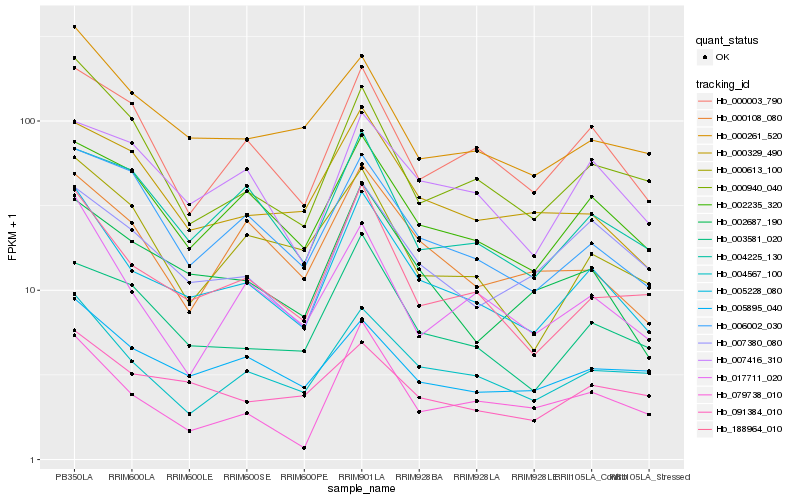
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005228_080 |
0.0 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 2 |
Hb_188964_010 |
0.0918730997 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 3 |
Hb_004567_100 |
0.1091860593 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 4 |
Hb_002687_190 |
0.1131296237 |
- |
- |
prolyl oligopeptidase [Cicer arietinum] |
| 5 |
Hb_002235_320 |
0.1146120433 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000003_790 |
0.1196367663 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 7 |
Hb_003581_020 |
0.1221182379 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000261_520 |
0.1243479087 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 9 |
Hb_000108_080 |
0.1282041883 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 10 |
Hb_000940_040 |
0.1282976796 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 11 |
Hb_006002_030 |
0.1286250542 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000329_490 |
0.1292761805 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 13 |
Hb_017711_020 |
0.1299495459 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007416_310 |
0.1310605284 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 15 |
Hb_005895_040 |
0.1314546336 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 16 |
Hb_007380_080 |
0.1315002656 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 17 |
Hb_091384_010 |
0.1320416038 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 18 |
Hb_000613_100 |
0.1328981547 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 19 |
Hb_004225_130 |
0.1329798078 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 20 |
Hb_079738_010 |
0.1336944092 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |