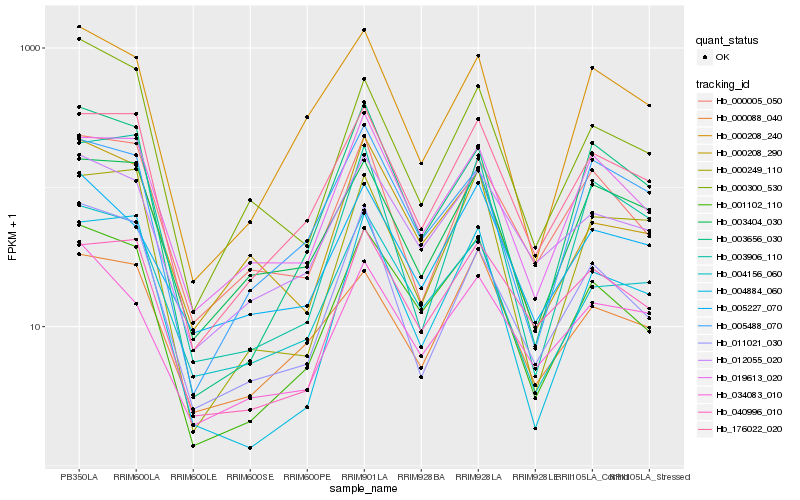
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_110 |
0.0 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1-like [Jatropha curcas] |
| 2 |
Hb_176022_020 |
0.1068882019 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_004156_060 |
0.1168703291 |
- |
- |
neutral/alkaline invertase 1 [Hevea brasiliensis] |
| 4 |
Hb_012055_020 |
0.1215474352 |
- |
- |
PREDICTED: metal transporter Nramp3 [Jatropha curcas] |
| 5 |
Hb_004884_060 |
0.1228613989 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 6 |
Hb_003656_030 |
0.1263747919 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 7 |
Hb_040996_010 |
0.1289623216 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase SEN1-like [Jatropha curcas] |
| 8 |
Hb_019613_020 |
0.1297338265 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 9 |
Hb_000208_290 |
0.1333682071 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000208_240 |
0.1336733192 |
- |
- |
PREDICTED: uncharacterized protein LOC105631307 [Jatropha curcas] |
| 11 |
Hb_000088_040 |
0.1372353396 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_011021_030 |
0.1398117698 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 13 |
Hb_000249_110 |
0.1438947641 |
- |
- |
hypothetical protein JCGZ_22462 [Jatropha curcas] |
| 14 |
Hb_005227_070 |
0.1468416598 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000300_530 |
0.1481746343 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0286299-like [Gossypium raimondii] |
| 16 |
Hb_005488_070 |
0.1504753176 |
- |
- |
putative cytochrome P450 [Oryza sativa Japonica Group] |
| 17 |
Hb_000005_050 |
0.1508631119 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 18 |
Hb_034083_010 |
0.1511038308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003404_030 |
0.154982349 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003906_110 |
0.1557934617 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase [Jatropha curcas] |