| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
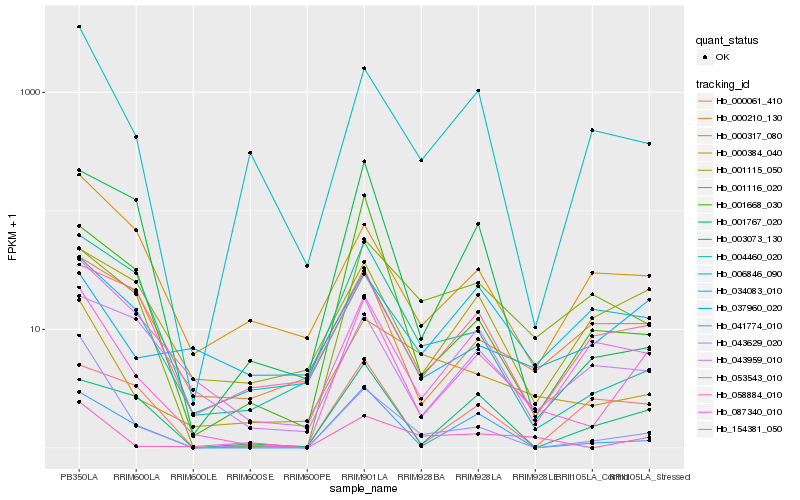
Hb_058884_010 |
0.0 |
- |
- |
PREDICTED: barwin-like [Jatropha curcas] |
| 2 |
Hb_041774_010 |
0.2171920564 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_003073_130 |
0.2450460341 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 4 |
Hb_087340_010 |
0.2468815512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_053543_010 |
0.2487694616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001668_030 |
0.2574966434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000384_040 |
0.2575579274 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 8 |
Hb_006846_090 |
0.2577575557 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 9 |
Hb_001115_050 |
0.2587891909 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 10 |
Hb_004460_020 |
0.2592814746 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 11 |
Hb_000210_130 |
0.261406589 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 12 |
Hb_154381_050 |
0.2656310505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000061_410 |
0.2709355272 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 14 |
Hb_034083_010 |
0.2723705843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_037960_020 |
0.2742276685 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 16 |
Hb_043959_010 |
0.2754974795 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X2 [Jatropha curcas] |
| 17 |
Hb_000317_080 |
0.2759047197 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 18 |
Hb_001767_020 |
0.2763661434 |
- |
- |
PREDICTED: WAT1-related protein At1g68170-like, partial [Jatropha curcas] |
| 19 |
Hb_043629_020 |
0.2777169021 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 20 |
Hb_001116_020 |
0.278978262 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2-like [Eucalyptus grandis] |