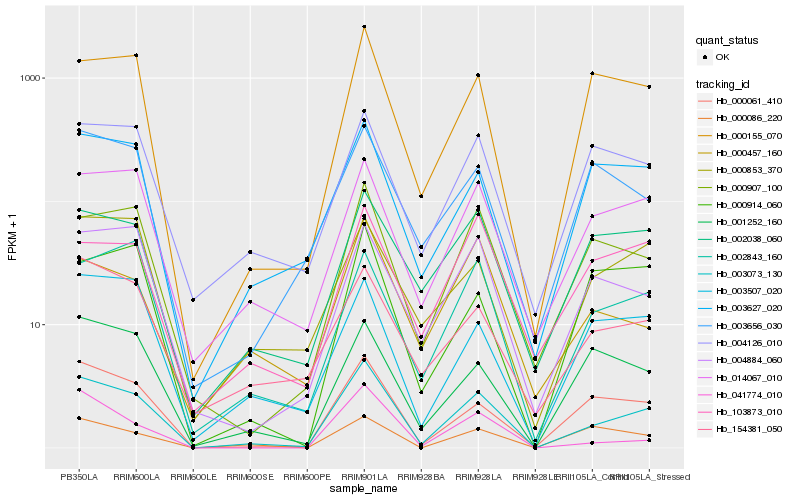
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_130 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 2 |
Hb_000061_410 |
0.1431508878 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000155_070 |
0.1598518782 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_014067_010 |
0.1650549699 |
- |
- |
hypothetical protein JCGZ_09524 [Jatropha curcas] |
| 5 |
Hb_000907_100 |
0.1685418212 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0012s03770g, partial [Populus trichocarpa] |
| 6 |
Hb_003627_020 |
0.1760815986 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 7 |
Hb_004884_060 |
0.1808963138 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 8 |
Hb_003507_020 |
0.1809332794 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 9 |
Hb_001252_160 |
0.1812189134 |
- |
- |
PREDICTED: PRA1 family protein A3 [Jatropha curcas] |
| 10 |
Hb_041774_010 |
0.1816043601 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 11 |
Hb_002843_160 |
0.1817133202 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Jatropha curcas] |
| 12 |
Hb_002038_060 |
0.1858596662 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100265010 [Vitis vinifera] |
| 13 |
Hb_154381_050 |
0.1861849419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004126_010 |
0.1892214786 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 15 |
Hb_000457_160 |
0.194161343 |
- |
- |
PREDICTED: uncharacterized protein LOC105633076 [Jatropha curcas] |
| 16 |
Hb_000853_370 |
0.1955581915 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 17 |
Hb_000914_060 |
0.1972048222 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 18 |
Hb_000086_220 |
0.1972248763 |
- |
- |
- |
| 19 |
Hb_003656_030 |
0.1987628092 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 20 |
Hb_103873_010 |
0.198879405 |
- |
- |
hypothetical protein POPTR_0008s14260g [Populus trichocarpa] |