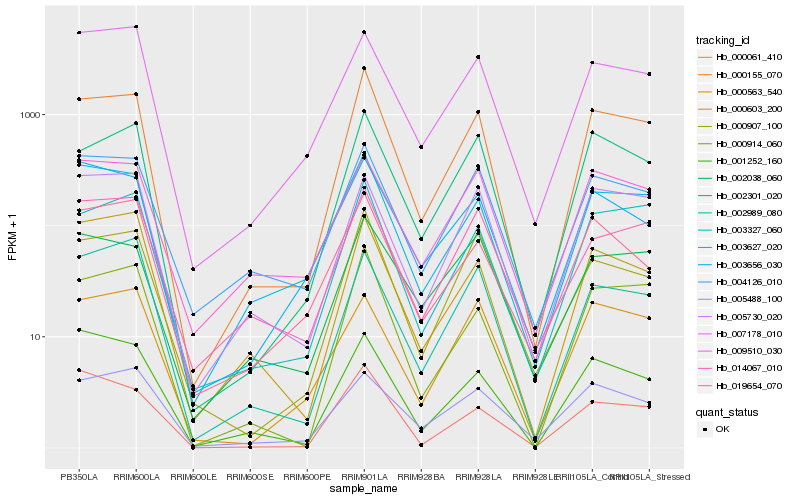
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_070 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 2 |
Hb_000914_060 |
0.0906771416 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 3 |
Hb_002301_020 |
0.0976832503 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_003327_060 |
0.1010422077 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003627_020 |
0.1037943692 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 6 |
Hb_000907_100 |
0.1084116617 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0012s03770g, partial [Populus trichocarpa] |
| 7 |
Hb_004126_010 |
0.1179919733 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 8 |
Hb_005730_020 |
0.121860829 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.1-like [Jatropha curcas] |
| 9 |
Hb_000061_410 |
0.1232564032 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_001252_160 |
0.1236026937 |
- |
- |
PREDICTED: PRA1 family protein A3 [Jatropha curcas] |
| 11 |
Hb_000603_200 |
0.1271931334 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 12 |
Hb_014067_010 |
0.1288909661 |
- |
- |
hypothetical protein JCGZ_09524 [Jatropha curcas] |
| 13 |
Hb_007178_010 |
0.1298598638 |
- |
- |
pseudo-hevein [Hevea brasiliensis] |
| 14 |
Hb_003656_030 |
0.1321605167 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 15 |
Hb_005488_100 |
0.1322760009 |
- |
- |
hypothetical protein JCGZ_00295 [Jatropha curcas] |
| 16 |
Hb_002989_080 |
0.1324398721 |
- |
- |
hypothetical protein JCGZ_22462 [Jatropha curcas] |
| 17 |
Hb_002038_060 |
0.1328385935 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100265010 [Vitis vinifera] |
| 18 |
Hb_019654_070 |
0.1366782627 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 19 |
Hb_000563_540 |
0.1378000269 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 20 |
Hb_009510_030 |
0.1384248968 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |