| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
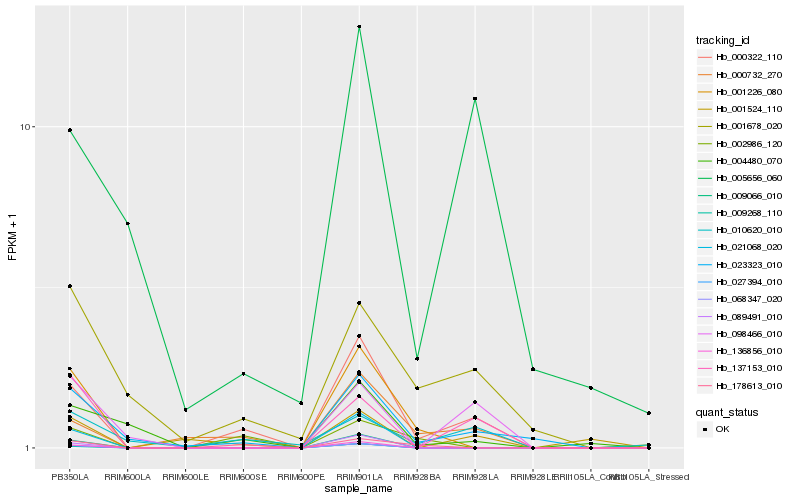
Hb_000322_110 |
0.0 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 3 [Jatropha curcas] |
| 2 |
Hb_000732_270 |
0.2104486347 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 3 |
Hb_023323_010 |
0.2529157099 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_010620_010 |
0.3075196884 |
- |
- |
hypothetical protein JCGZ_22570 [Jatropha curcas] |
| 5 |
Hb_178613_010 |
0.3136600185 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 6 |
Hb_004480_070 |
0.3181047069 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_137153_010 |
0.3190419127 |
- |
- |
- |
| 8 |
Hb_001226_080 |
0.3209771708 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_002986_120 |
0.3242121914 |
- |
- |
- |
| 10 |
Hb_001678_020 |
0.3248113891 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 11 |
Hb_005656_060 |
0.3271695493 |
- |
- |
- |
| 12 |
Hb_136856_010 |
0.3295466756 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 13 |
Hb_021068_020 |
0.3297837093 |
- |
- |
- |
| 14 |
Hb_009066_010 |
0.3311456172 |
- |
- |
- |
| 15 |
Hb_068347_020 |
0.331654202 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_027394_010 |
0.331656471 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 17 |
Hb_089491_010 |
0.3320846274 |
- |
- |
- |
| 18 |
Hb_098466_010 |
0.3322463465 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 19 |
Hb_009268_110 |
0.3323128195 |
- |
- |
PREDICTED: uncharacterized protein LOC102611294 [Citrus sinensis] |
| 20 |
Hb_001524_110 |
0.3337284411 |
- |
- |
PREDICTED: uncharacterized protein LOC104611877 isoform X2 [Nelumbo nucifera] |