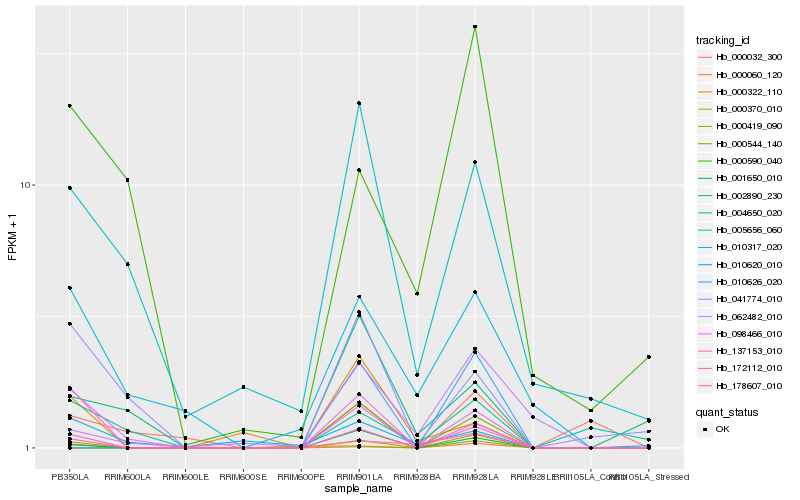
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172112_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 2 |
Hb_001650_010 |
0.2187156191 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_098466_010 |
0.2303456489 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 4 |
Hb_137153_010 |
0.2549671289 |
- |
- |
- |
| 5 |
Hb_178607_010 |
0.2586142504 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 6 |
Hb_005656_060 |
0.3209181014 |
- |
- |
- |
| 7 |
Hb_000060_120 |
0.3213532629 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 8 |
Hb_000419_090 |
0.3298636306 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 9 |
Hb_041774_010 |
0.336639409 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 10 |
Hb_010317_020 |
0.342364675 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 11 |
Hb_000322_110 |
0.3537428822 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 3 [Jatropha curcas] |
| 12 |
Hb_062482_010 |
0.3632890453 |
- |
- |
- |
| 13 |
Hb_000590_040 |
0.3659728787 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 14 |
Hb_010626_020 |
0.3681396899 |
- |
- |
- |
| 15 |
Hb_002890_230 |
0.3789232937 |
- |
- |
hypothetical protein MTR_5g084920 [Medicago truncatula] |
| 16 |
Hb_000370_010 |
0.3791753004 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000032_300 |
0.3800417463 |
- |
- |
hypothetical protein JCGZ_20455 [Jatropha curcas] |
| 18 |
Hb_010620_010 |
0.3810629377 |
- |
- |
hypothetical protein JCGZ_22570 [Jatropha curcas] |
| 19 |
Hb_000544_140 |
0.3832410559 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 20 |
Hb_004650_020 |
0.3846254594 |
- |
- |
- |