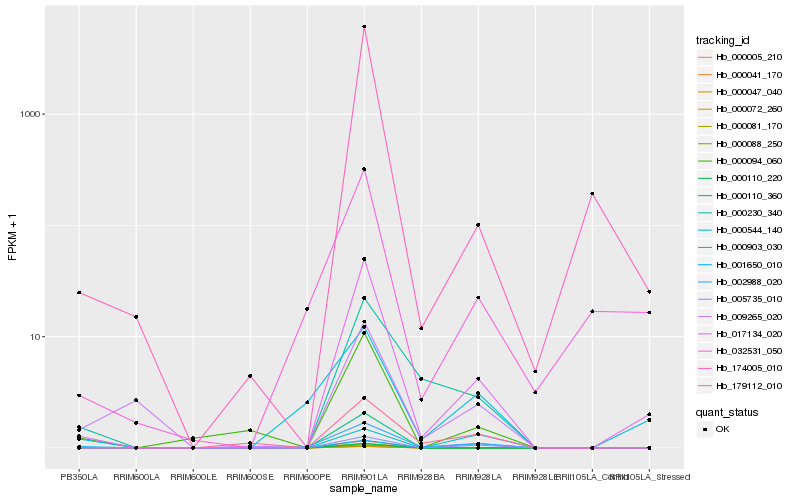
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005735_010 |
0.0 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002988_020 |
0.1816949298 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 3 |
Hb_017134_020 |
0.1969734166 |
- |
- |
- |
| 4 |
Hb_000544_140 |
0.2043365978 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 5 |
Hb_179112_010 |
0.2135708502 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 6 |
Hb_000094_060 |
0.2636533388 |
- |
- |
- |
| 7 |
Hb_001650_010 |
0.2778665953 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_174005_010 |
0.287978069 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 9 |
Hb_009265_020 |
0.2897268972 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000230_340 |
0.2965331336 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 11 |
Hb_000903_030 |
0.3020529081 |
- |
- |
- |
| 12 |
Hb_032531_050 |
0.3080232101 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 13 |
Hb_000005_210 |
0.3106257802 |
- |
- |
- |
| 14 |
Hb_000041_170 |
0.3106257802 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 15 |
Hb_000047_040 |
0.3106257802 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 16 |
Hb_000072_260 |
0.3106257802 |
- |
- |
hypothetical protein MTR_5g044090 [Medicago truncatula] |
| 17 |
Hb_000081_170 |
0.3106257802 |
- |
- |
- |
| 18 |
Hb_000088_250 |
0.3106257802 |
- |
- |
- |
| 19 |
Hb_000110_220 |
0.3106257802 |
- |
- |
- |
| 20 |
Hb_000110_360 |
0.3106257802 |
- |
- |
- |