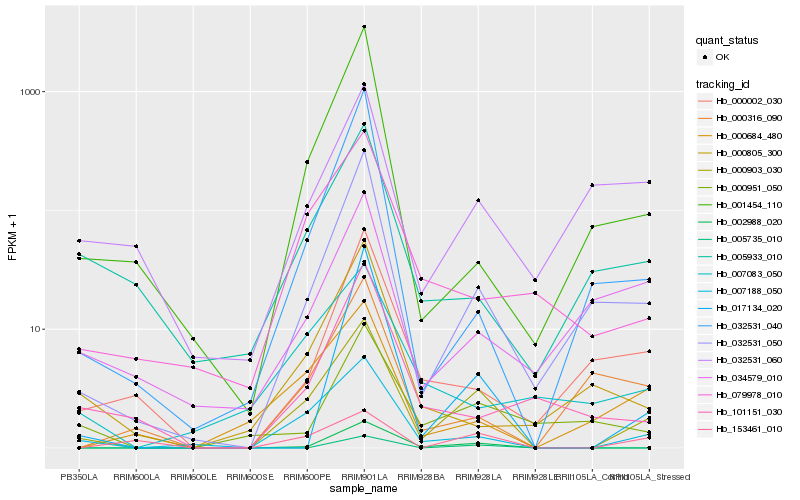
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000903_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_007188_050 |
0.1763224573 |
- |
- |
- |
| 3 |
Hb_032531_050 |
0.2215255244 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 4 |
Hb_002988_020 |
0.2245911175 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 5 |
Hb_153461_010 |
0.2653749927 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 6 |
Hb_079978_010 |
0.2660463768 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 7 |
Hb_000684_480 |
0.2716617886 |
transcription factor |
TF Family: Orphans |
- |
| 8 |
Hb_017134_020 |
0.2752008819 |
- |
- |
- |
| 9 |
Hb_001454_110 |
0.2754844538 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 10 |
Hb_032531_040 |
0.2782377309 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 11 |
Hb_005933_010 |
0.2885758476 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 12 |
Hb_000002_030 |
0.2926776464 |
- |
- |
- |
| 13 |
Hb_000951_050 |
0.2926901588 |
- |
- |
- |
| 14 |
Hb_032531_060 |
0.2942043897 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 15 |
Hb_000805_300 |
0.2942822567 |
- |
- |
- |
| 16 |
Hb_007083_050 |
0.2967414018 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 17 |
Hb_000316_090 |
0.3005547802 |
- |
- |
- |
| 18 |
Hb_101151_030 |
0.3005733739 |
- |
- |
- |
| 19 |
Hb_005735_010 |
0.3020529081 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_034579_010 |
0.3037807247 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |