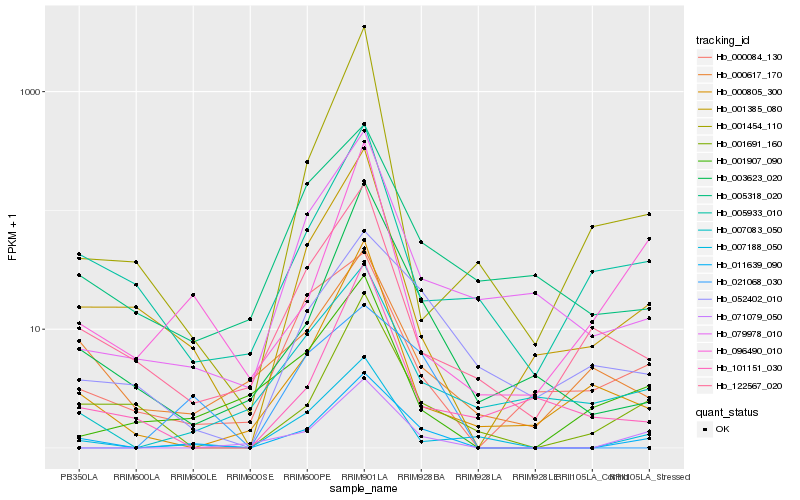
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011639_090 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH75-like [Populus euphratica] |
| 2 |
Hb_007188_050 |
0.2140161189 |
- |
- |
- |
| 3 |
Hb_003623_020 |
0.2173694483 |
- |
- |
- |
| 4 |
Hb_071079_050 |
0.229240427 |
- |
- |
- |
| 5 |
Hb_001385_080 |
0.229395535 |
- |
- |
- |
| 6 |
Hb_001691_160 |
0.2415268381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_079978_010 |
0.2578594154 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_007083_050 |
0.2597110246 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_122567_020 |
0.2597215823 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 10 |
Hb_000805_300 |
0.2642372486 |
- |
- |
- |
| 11 |
Hb_052402_010 |
0.267487276 |
- |
- |
- |
| 12 |
Hb_005933_010 |
0.268939543 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 13 |
Hb_001907_090 |
0.2722734402 |
- |
- |
- |
| 14 |
Hb_005318_020 |
0.2733760677 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000617_170 |
0.2752412047 |
- |
- |
- |
| 16 |
Hb_000084_130 |
0.2792990689 |
- |
- |
- |
| 17 |
Hb_096490_010 |
0.2802279725 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 18 |
Hb_101151_030 |
0.2880544631 |
- |
- |
- |
| 19 |
Hb_021068_030 |
0.2898813671 |
transcription factor |
TF Family: M-type |
hypothetical protein B456_010G032700 [Gossypium raimondii] |
| 20 |
Hb_001454_110 |
0.2905249814 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |