| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
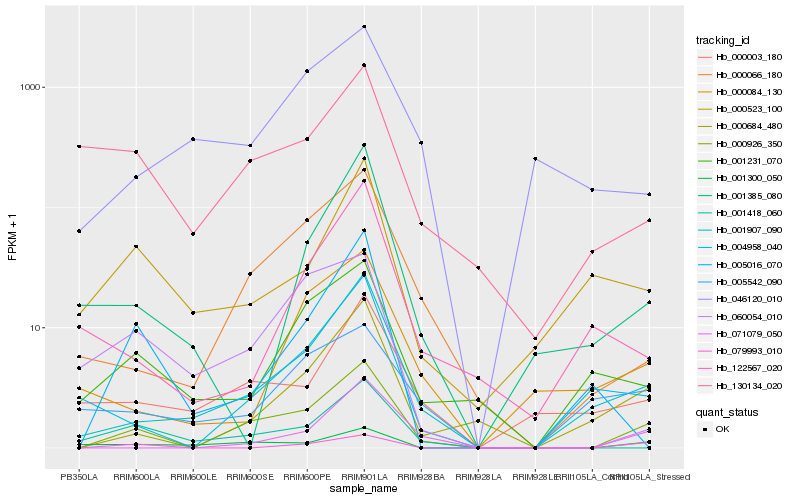
Hb_000926_350 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: upstream activation factor subunit UAF30 [Jatropha curcas] |
| 2 |
Hb_001907_090 |
0.2193109111 |
- |
- |
- |
| 3 |
Hb_000684_480 |
0.2382696036 |
transcription factor |
TF Family: Orphans |
- |
| 4 |
Hb_000066_180 |
0.2473837977 |
- |
- |
hypothetical protein POPTR_0008s02710g [Populus trichocarpa] |
| 5 |
Hb_071079_050 |
0.2546755952 |
- |
- |
- |
| 6 |
Hb_004958_040 |
0.2721814867 |
- |
- |
- |
| 7 |
Hb_001300_050 |
0.2883781673 |
- |
- |
- |
| 8 |
Hb_001418_060 |
0.2915219472 |
- |
- |
- |
| 9 |
Hb_079993_010 |
0.293601961 |
- |
- |
PREDICTED: uncharacterized protein LOC103336399 [Prunus mume] |
| 10 |
Hb_000003_180 |
0.2943212363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001231_070 |
0.2986752573 |
- |
- |
- |
| 12 |
Hb_130134_020 |
0.3073552033 |
- |
- |
- |
| 13 |
Hb_000523_100 |
0.3128228263 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 14 |
Hb_000084_130 |
0.3135671116 |
- |
- |
- |
| 15 |
Hb_005016_070 |
0.3145865742 |
- |
- |
- |
| 16 |
Hb_001385_080 |
0.3165799699 |
- |
- |
- |
| 17 |
Hb_060054_010 |
0.3178648447 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 18 |
Hb_046120_010 |
0.3191716334 |
- |
- |
- |
| 19 |
Hb_005542_090 |
0.3213997259 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 20 |
Hb_122567_020 |
0.3249641665 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |