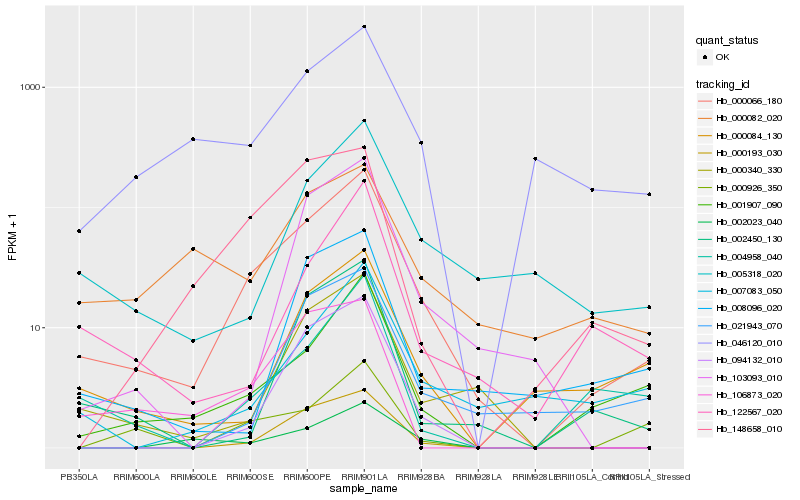
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000066_180 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s02710g [Populus trichocarpa] |
| 2 |
Hb_001907_090 |
0.198334255 |
- |
- |
- |
| 3 |
Hb_000193_030 |
0.2110575782 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 4 |
Hb_000340_330 |
0.2147586416 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 5 |
Hb_005318_020 |
0.2177403559 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_148658_010 |
0.2239942688 |
- |
- |
- |
| 7 |
Hb_094132_010 |
0.2242565096 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 8 |
Hb_002450_130 |
0.2277334715 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 9 |
Hb_046120_010 |
0.2294238922 |
- |
- |
- |
| 10 |
Hb_007083_050 |
0.2335205956 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 11 |
Hb_004958_040 |
0.233954187 |
- |
- |
- |
| 12 |
Hb_122567_020 |
0.2370193139 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 13 |
Hb_000084_130 |
0.237264714 |
- |
- |
- |
| 14 |
Hb_002023_040 |
0.2388101568 |
- |
- |
- |
| 15 |
Hb_103093_010 |
0.2398322111 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 16 |
Hb_021943_070 |
0.2438935697 |
- |
- |
- |
| 17 |
Hb_008096_020 |
0.246419456 |
- |
- |
- |
| 18 |
Hb_000926_350 |
0.2473837977 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: upstream activation factor subunit UAF30 [Jatropha curcas] |
| 19 |
Hb_000082_020 |
0.2492533813 |
- |
- |
- |
| 20 |
Hb_106873_020 |
0.250049068 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |