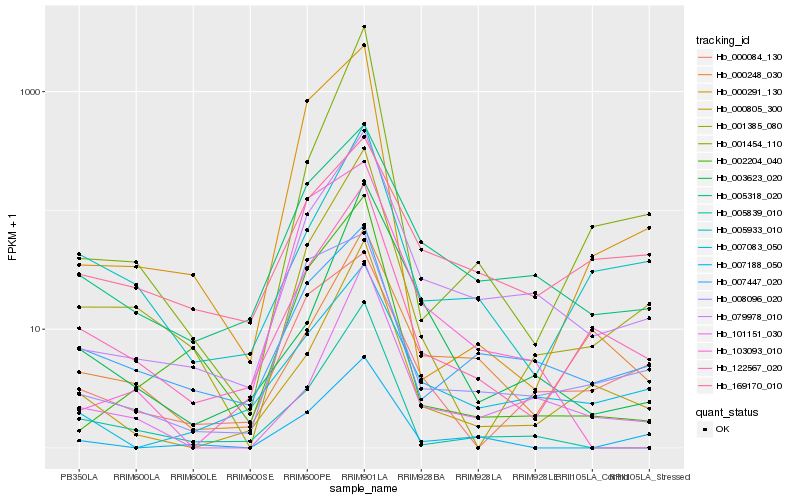
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079978_010 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 2 |
Hb_005318_020 |
0.1265110485 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 3 |
Hb_007083_050 |
0.1313016216 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_122567_020 |
0.1677401923 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 5 |
Hb_101151_030 |
0.1713519793 |
- |
- |
- |
| 6 |
Hb_000805_300 |
0.1773622767 |
- |
- |
- |
| 7 |
Hb_007447_020 |
0.1833666544 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 8 |
Hb_001385_080 |
0.1898489953 |
- |
- |
- |
| 9 |
Hb_007188_050 |
0.190501523 |
- |
- |
- |
| 10 |
Hb_003623_020 |
0.1934994735 |
- |
- |
- |
| 11 |
Hb_002204_040 |
0.1935943516 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 12 |
Hb_008096_020 |
0.2071644093 |
- |
- |
- |
| 13 |
Hb_000084_130 |
0.2072457093 |
- |
- |
- |
| 14 |
Hb_000248_030 |
0.207293143 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 15 |
Hb_169170_010 |
0.2081689146 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_005933_010 |
0.208835487 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 17 |
Hb_005839_010 |
0.2125740937 |
- |
- |
30S ribosomal protein s13 [Camellia sinensis] |
| 18 |
Hb_001454_110 |
0.2156410219 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 19 |
Hb_103093_010 |
0.2187774338 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 20 |
Hb_000291_130 |
0.2206960052 |
- |
- |
- |