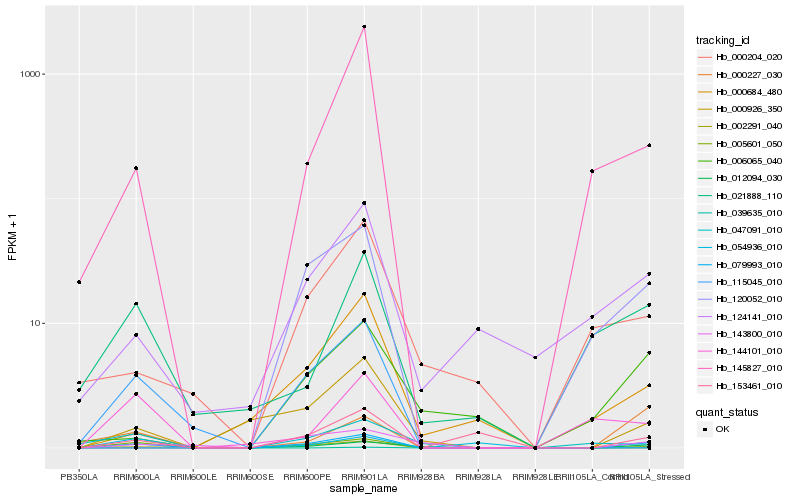
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079993_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103336399 [Prunus mume] |
| 2 |
Hb_002291_040 |
0.2656241042 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 3 |
Hb_000926_350 |
0.293601961 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: upstream activation factor subunit UAF30 [Jatropha curcas] |
| 4 |
Hb_145827_010 |
0.2973610183 |
- |
- |
hypothetical protein POPTR_0003s13630g [Populus trichocarpa] |
| 5 |
Hb_000227_030 |
0.3076217763 |
- |
- |
- |
| 6 |
Hb_153461_010 |
0.3127064361 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 7 |
Hb_124141_010 |
0.3247238388 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 8 |
Hb_039635_010 |
0.3254779205 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 9 |
Hb_143800_010 |
0.3258632234 |
- |
- |
hypothetical protein POPTR_0010s21410g, partial [Populus trichocarpa] |
| 10 |
Hb_120052_010 |
0.3295801967 |
- |
- |
hypothetical protein PRUPE_ppa1027171mg [Prunus persica] |
| 11 |
Hb_054936_010 |
0.3330976353 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 12 |
Hb_000684_480 |
0.3355154053 |
transcription factor |
TF Family: Orphans |
- |
| 13 |
Hb_021888_110 |
0.3381497756 |
- |
- |
- |
| 14 |
Hb_006065_040 |
0.3387176186 |
- |
- |
- |
| 15 |
Hb_144101_010 |
0.3468971062 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 16 |
Hb_012094_030 |
0.3486044924 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 17 |
Hb_115045_010 |
0.3487899375 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_005601_050 |
0.3492574529 |
- |
- |
PREDICTED: uncharacterized protein LOC103444007 [Malus domestica] |
| 19 |
Hb_047091_010 |
0.3536423311 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 20 |
Hb_000204_020 |
0.355006118 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |