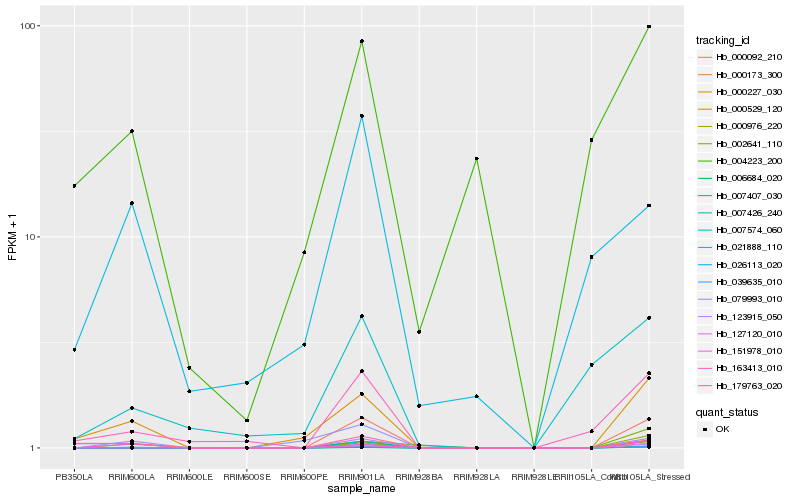
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_039635_010 |
0.2377575906 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_151978_010 |
0.2554793843 |
- |
- |
- |
| 4 |
Hb_179763_020 |
0.2607153974 |
- |
- |
- |
| 5 |
Hb_079993_010 |
0.3076217763 |
- |
- |
PREDICTED: uncharacterized protein LOC103336399 [Prunus mume] |
| 6 |
Hb_163413_010 |
0.3088872336 |
- |
- |
PREDICTED: uncharacterized protein LOC102604542 [Solanum tuberosum] |
| 7 |
Hb_002641_110 |
0.3245145136 |
- |
- |
PREDICTED: uncharacterized protein LOC103448574 [Malus domestica] |
| 8 |
Hb_004223_200 |
0.3245474267 |
- |
- |
PREDICTED: auxin-induced protein 6B-like [Cucumis sativus] |
| 9 |
Hb_123915_050 |
0.3383873974 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 10 |
Hb_000529_120 |
0.3504465689 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 11 |
Hb_007426_240 |
0.3521491727 |
- |
- |
- |
| 12 |
Hb_000173_300 |
0.3530118622 |
- |
- |
PREDICTED: uncharacterized protein LOC105631894 [Jatropha curcas] |
| 13 |
Hb_026113_020 |
0.3544799091 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_006684_020 |
0.3549626981 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_000976_220 |
0.3568185938 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4-like [Jatropha curcas] |
| 16 |
Hb_127120_010 |
0.3568283595 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 17 |
Hb_007407_030 |
0.3568645037 |
- |
- |
hypothetical protein JCGZ_23510 [Jatropha curcas] |
| 18 |
Hb_000092_210 |
0.3589279729 |
- |
- |
- |
| 19 |
Hb_021888_110 |
0.3623263635 |
- |
- |
- |
| 20 |
Hb_007574_060 |
0.3627838149 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 27-like [Jatropha curcas] |