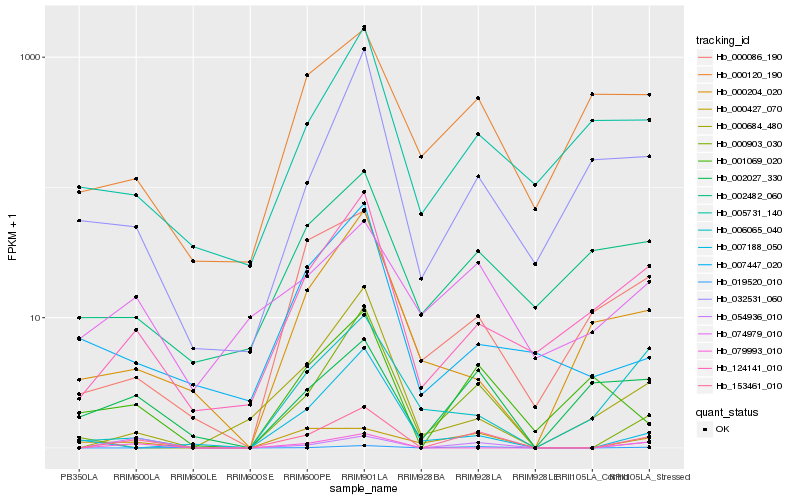
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153461_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 2 |
Hb_054936_010 |
0.2552405436 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 3 |
Hb_000903_030 |
0.2653749927 |
- |
- |
- |
| 4 |
Hb_124141_010 |
0.2967503174 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 5 |
Hb_000684_480 |
0.3103847658 |
transcription factor |
TF Family: Orphans |
- |
| 6 |
Hb_079993_010 |
0.3127064361 |
- |
- |
PREDICTED: uncharacterized protein LOC103336399 [Prunus mume] |
| 7 |
Hb_000086_190 |
0.3287017493 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 8 |
Hb_001069_020 |
0.3312659037 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_032531_060 |
0.3318207378 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 10 |
Hb_002027_330 |
0.3320300791 |
- |
- |
- |
| 11 |
Hb_000427_070 |
0.3331981918 |
- |
- |
- |
| 12 |
Hb_019520_010 |
0.3390772211 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 13 |
Hb_006065_040 |
0.3406344541 |
- |
- |
- |
| 14 |
Hb_007188_050 |
0.3419639746 |
- |
- |
- |
| 15 |
Hb_005731_140 |
0.3467988798 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 16 |
Hb_000204_020 |
0.3550671433 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 17 |
Hb_000120_190 |
0.3558064201 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 18 |
Hb_074979_010 |
0.3567201839 |
- |
- |
hypothetical protein AALP_AA2G213100 [Arabis alpina] |
| 19 |
Hb_007447_020 |
0.3567289385 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 20 |
Hb_002482_060 |
0.357364647 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |