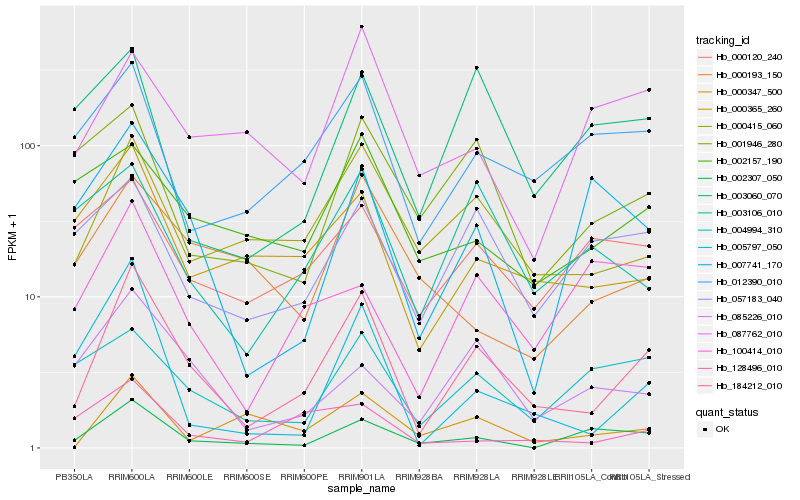
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184212_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105113658 [Populus euphratica] |
| 2 |
Hb_000415_060 |
0.216003266 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 3 |
Hb_005797_050 |
0.2385840871 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 4 |
Hb_002307_050 |
0.2490038454 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000365_260 |
0.2491308732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012390_010 |
0.2513590169 |
- |
- |
pectate lyase 1 [Hevea brasiliensis] |
| 7 |
Hb_085226_010 |
0.2580035094 |
- |
- |
hypothetical protein POPTR_0002s03930g [Populus trichocarpa] |
| 8 |
Hb_057183_040 |
0.2637647437 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 2 [Jatropha curcas] |
| 9 |
Hb_000347_500 |
0.2668033762 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001946_280 |
0.2690200529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004994_310 |
0.2697145726 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 12 |
Hb_087762_010 |
0.2748220386 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 13 |
Hb_003106_010 |
0.2753790229 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 14 |
Hb_128496_010 |
0.2757886991 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_002157_190 |
0.2758706907 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 16 |
Hb_007741_170 |
0.2795806964 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4-like [Jatropha curcas] |
| 17 |
Hb_100414_010 |
0.2796501812 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_000193_150 |
0.2816996597 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 19 |
Hb_000120_240 |
0.2819034462 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 20 |
Hb_003060_070 |
0.2829556649 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |