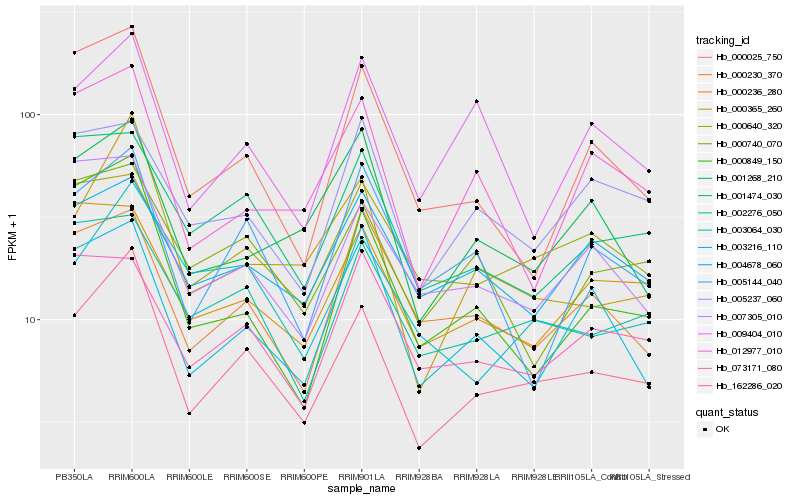
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162286_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642702 [Jatropha curcas] |
| 2 |
Hb_000365_260 |
0.1188952318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002276_050 |
0.128697044 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001474_030 |
0.1389344152 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 5 |
Hb_007305_010 |
0.1431774156 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 6 |
Hb_003064_030 |
0.1443473262 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_004678_060 |
0.1457522386 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 8 |
Hb_003216_110 |
0.1459761798 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 9 |
Hb_000025_750 |
0.1469916853 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 10 |
Hb_000640_320 |
0.1479450738 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000236_280 |
0.1483544817 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000849_150 |
0.1484343294 |
- |
- |
hypothetical protein JCGZ_15099 [Jatropha curcas] |
| 13 |
Hb_000230_370 |
0.1488590873 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_012977_010 |
0.1491864139 |
- |
- |
PREDICTED: uncharacterized protein LOC105131558 [Populus euphratica] |
| 15 |
Hb_001268_210 |
0.1507630801 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000740_070 |
0.1510162523 |
- |
- |
- |
| 17 |
Hb_005237_060 |
0.1512035927 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 18 |
Hb_073171_080 |
0.1520650492 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_009404_010 |
0.1520783144 |
- |
- |
PREDICTED: casein kinase II subunit alpha-2-like [Glycine max] |
| 20 |
Hb_005144_040 |
0.1526117762 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |