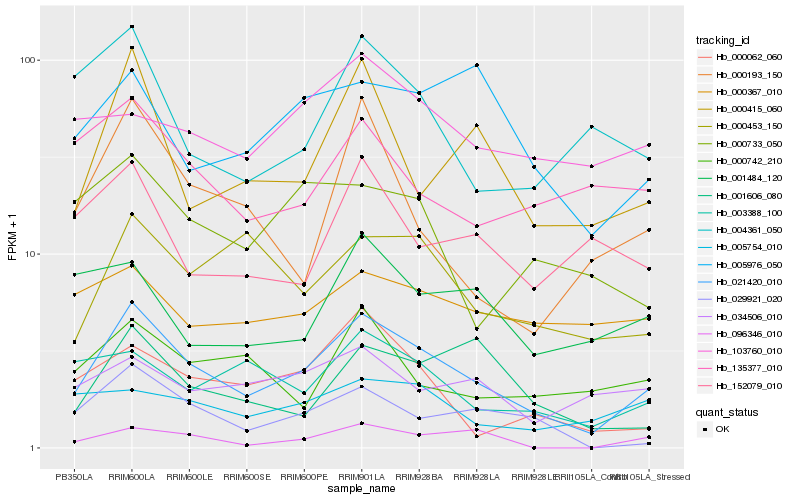
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021420_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10029508mg [Citrus clementina] |
| 2 |
Hb_000415_060 |
0.187922881 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 3 |
Hb_029921_020 |
0.197883404 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000453_150 |
0.1985921952 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 5 |
Hb_003388_100 |
0.2028561431 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 6 |
Hb_000062_060 |
0.2040997266 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 7 |
Hb_000193_150 |
0.2066779238 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 8 |
Hb_004361_050 |
0.2086320445 |
- |
- |
ring finger containing protein, putative [Ricinus communis] |
| 9 |
Hb_005754_010 |
0.2102835808 |
- |
- |
hypothetical protein POPTR_0660s00210g [Populus trichocarpa] |
| 10 |
Hb_001606_080 |
0.21093473 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005976_050 |
0.2113017096 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 12 |
Hb_034506_010 |
0.2124247432 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 13 |
Hb_000742_210 |
0.2131268503 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_096346_010 |
0.2141318833 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000733_050 |
0.2144660934 |
- |
- |
PREDICTED: uncharacterized protein LOC105646910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001484_120 |
0.2171258061 |
- |
- |
hypothetical protein POPTR_2287s00200g, partial [Populus trichocarpa] |
| 17 |
Hb_135377_010 |
0.2173474147 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 18 |
Hb_152079_010 |
0.2178557663 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 19 |
Hb_103760_010 |
0.2186130243 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 20 |
Hb_000367_010 |
0.2205222737 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |