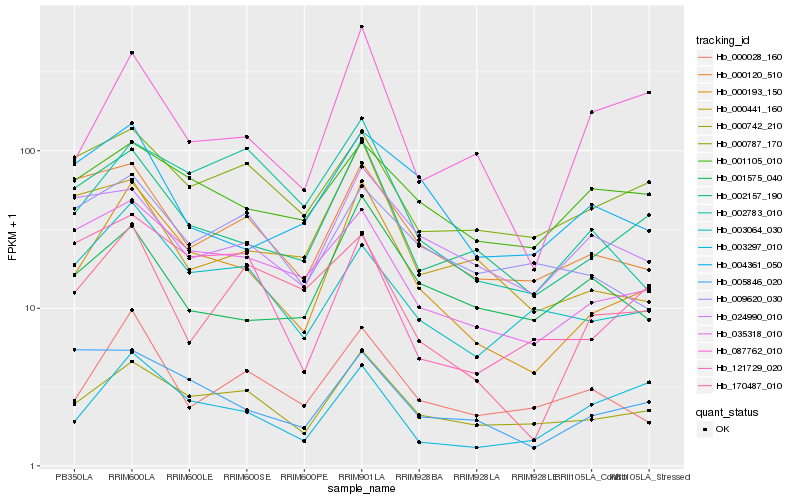
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000193_150 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 2 |
Hb_000742_210 |
0.1429281397 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_002157_190 |
0.1455354344 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 4 |
Hb_035318_010 |
0.1651792103 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 5 |
Hb_000028_160 |
0.1688808589 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 6 |
Hb_003064_030 |
0.178101934 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_087762_010 |
0.1798393274 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 8 |
Hb_003297_010 |
0.1841981673 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 9 |
Hb_000120_510 |
0.1845490983 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002783_010 |
0.1900457288 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 11 |
Hb_001575_040 |
0.1921720677 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000787_170 |
0.1948038152 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 13 |
Hb_121729_020 |
0.1957434317 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000441_160 |
0.1973698912 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 15 |
Hb_004361_050 |
0.1984998717 |
- |
- |
ring finger containing protein, putative [Ricinus communis] |
| 16 |
Hb_170487_010 |
0.1987404253 |
- |
- |
- |
| 17 |
Hb_001105_010 |
0.1994701677 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_005846_020 |
0.1999889416 |
- |
- |
PREDICTED: tubulin-folding cofactor C [Jatropha curcas] |
| 19 |
Hb_009620_030 |
0.2060616202 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 20 |
Hb_024990_010 |
0.2062984893 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |