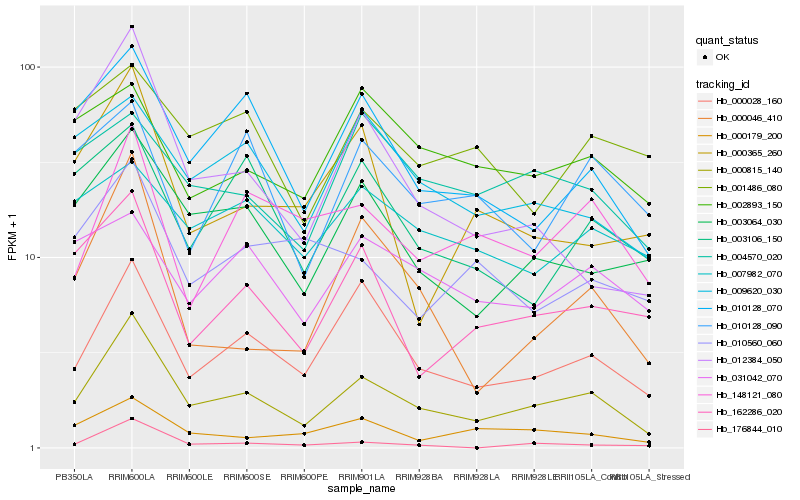
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_140 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000028_160 |
0.1401384378 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 3 |
Hb_003064_030 |
0.1623890748 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_010128_070 |
0.1638795278 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000046_410 |
0.17436014 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_162286_020 |
0.1763091162 |
- |
- |
PREDICTED: uncharacterized protein LOC105642702 [Jatropha curcas] |
| 7 |
Hb_012384_050 |
0.1808726077 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 8 |
Hb_000179_200 |
0.1818696118 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_148121_080 |
0.1874782461 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 10 |
Hb_009620_030 |
0.1910428051 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 11 |
Hb_176844_010 |
0.1957960304 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003106_150 |
0.1989979165 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 13 |
Hb_000365_260 |
0.2002132018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004570_020 |
0.2080403789 |
- |
- |
PREDICTED: protein TIC 100 [Jatropha curcas] |
| 15 |
Hb_031042_070 |
0.2087710433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010128_090 |
0.2109766902 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 17 |
Hb_010560_060 |
0.2117644318 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 18 |
Hb_007982_070 |
0.2129557712 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 19 |
Hb_002893_150 |
0.214197216 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 20 |
Hb_001486_080 |
0.2147501287 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |