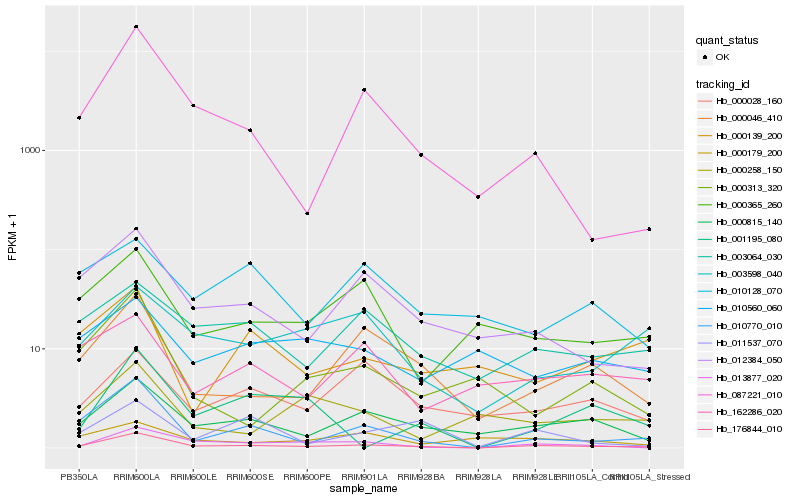
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176844_010 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_010770_010 |
0.1754271534 |
- |
- |
- |
| 3 |
Hb_000815_140 |
0.1957960304 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000046_410 |
0.2155780277 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_013877_020 |
0.2163567405 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 6 |
Hb_012384_050 |
0.2225387242 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 7 |
Hb_087221_010 |
0.2299647503 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 8 |
Hb_003064_030 |
0.239116797 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000365_260 |
0.2590681271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000028_160 |
0.2601282643 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 11 |
Hb_003598_040 |
0.2632602243 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 12 |
Hb_162286_020 |
0.2670875876 |
- |
- |
PREDICTED: uncharacterized protein LOC105642702 [Jatropha curcas] |
| 13 |
Hb_011537_070 |
0.2722705281 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 14 |
Hb_000313_320 |
0.2785565626 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 15 |
Hb_001195_080 |
0.2811116038 |
- |
- |
PREDICTED: uncharacterized protein LOC105633814 [Jatropha curcas] |
| 16 |
Hb_000139_200 |
0.2825042926 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_010560_060 |
0.2906606288 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 18 |
Hb_010128_070 |
0.2911326352 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000258_150 |
0.2914640998 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 20 |
Hb_000179_200 |
0.2928578134 |
- |
- |
unnamed protein product [Vitis vinifera] |