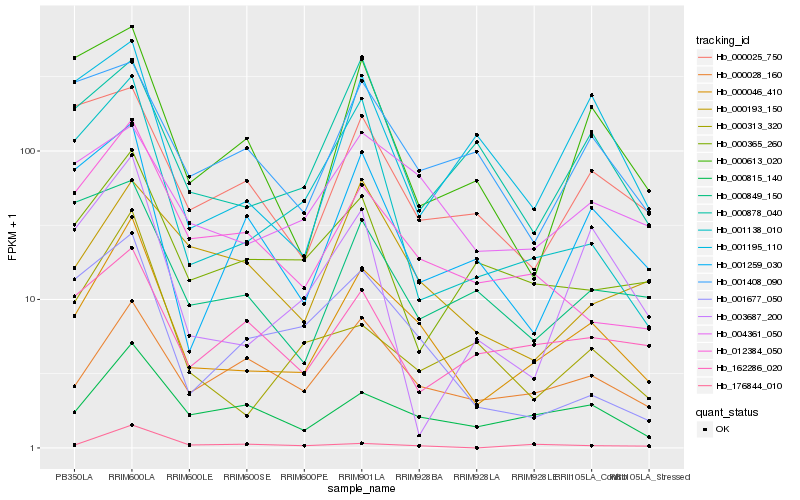
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_410 |
0.0 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_000815_140 |
0.17436014 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_012384_050 |
0.1933056784 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 4 |
Hb_000028_160 |
0.1945030326 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 5 |
Hb_004361_050 |
0.1983847066 |
- |
- |
ring finger containing protein, putative [Ricinus communis] |
| 6 |
Hb_176844_010 |
0.2155780277 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001138_010 |
0.2172191243 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_001259_030 |
0.2187719214 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 9 |
Hb_001677_050 |
0.224793104 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP8, putative [Ricinus communis] |
| 10 |
Hb_000365_260 |
0.2270952733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000313_320 |
0.229894856 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 12 |
Hb_003687_200 |
0.2300715451 |
- |
- |
PREDICTED: uncharacterized protein LOC105632139 [Jatropha curcas] |
| 13 |
Hb_000878_040 |
0.230082254 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000025_750 |
0.2304308933 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 15 |
Hb_162286_020 |
0.2311960491 |
- |
- |
PREDICTED: uncharacterized protein LOC105642702 [Jatropha curcas] |
| 16 |
Hb_000613_020 |
0.2328401883 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 17 |
Hb_000193_150 |
0.2348621249 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 18 |
Hb_000849_150 |
0.2360437229 |
- |
- |
hypothetical protein JCGZ_15099 [Jatropha curcas] |
| 19 |
Hb_001408_090 |
0.2370642915 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 20 |
Hb_001195_110 |
0.2404469341 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |