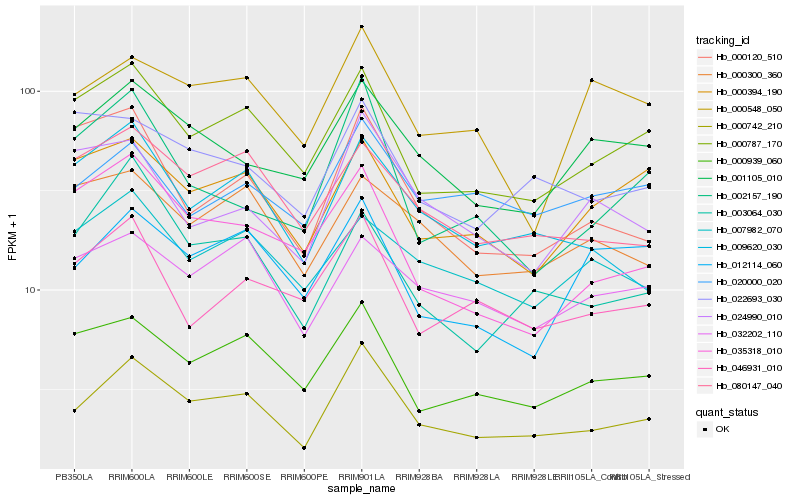
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_210 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_000787_170 |
0.1043348413 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 3 |
Hb_000939_060 |
0.1094890081 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 4 |
Hb_001105_010 |
0.123692954 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_080147_040 |
0.124590627 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 6 |
Hb_009620_030 |
0.1266032725 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 7 |
Hb_046931_010 |
0.1286456416 |
- |
- |
hypothetical protein JCGZ_26924 [Jatropha curcas] |
| 8 |
Hb_032202_110 |
0.1306050009 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 9 |
Hb_020000_020 |
0.1310014825 |
- |
- |
Troponin C, skeletal muscle, putative [Ricinus communis] |
| 10 |
Hb_012114_060 |
0.1324157394 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 11 |
Hb_000394_190 |
0.1324367439 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000120_510 |
0.1342543365 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002157_190 |
0.1357616757 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 14 |
Hb_024990_010 |
0.1364382892 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 15 |
Hb_035318_010 |
0.1364795898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 16 |
Hb_000300_360 |
0.1370280223 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 17 |
Hb_000548_050 |
0.1373799495 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 18 |
Hb_022693_030 |
0.137397719 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 19 |
Hb_007982_070 |
0.1383560807 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 20 |
Hb_003064_030 |
0.1386420151 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |