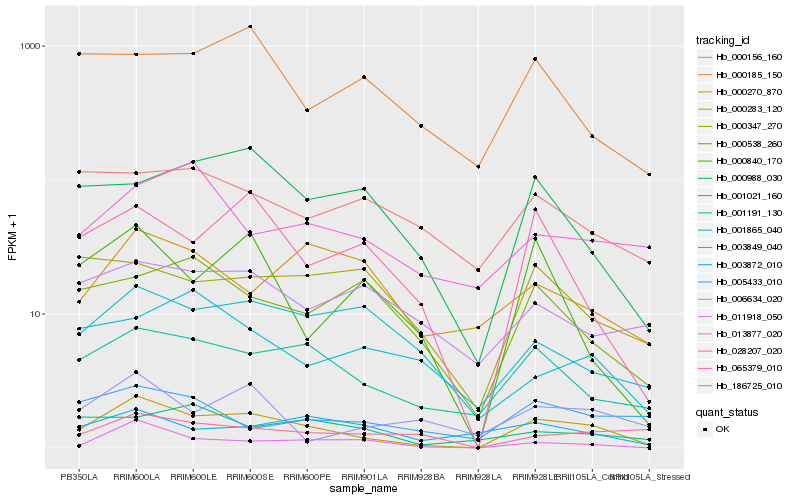
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 2 |
Hb_001191_130 |
0.2044079106 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic [Jatropha curcas] |
| 3 |
Hb_065379_010 |
0.2303718166 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 4 |
Hb_013877_020 |
0.2434541995 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 5 |
Hb_000988_030 |
0.2495718797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000840_170 |
0.2519928313 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 7 |
Hb_186725_010 |
0.2618405748 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_006634_020 |
0.2635512158 |
- |
- |
- |
| 9 |
Hb_000538_260 |
0.2655075093 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 10 |
Hb_000185_150 |
0.2659668221 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000347_270 |
0.270735988 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 12 |
Hb_001865_040 |
0.2708894764 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_003872_010 |
0.271458261 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 14 |
Hb_011918_050 |
0.2743467262 |
- |
- |
- |
| 15 |
Hb_005433_010 |
0.2749541295 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 16 |
Hb_000270_870 |
0.2756681487 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 17 |
Hb_003849_040 |
0.277617771 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 18 |
Hb_028207_020 |
0.2780445736 |
- |
- |
PREDICTED: uncharacterized protein LOC105650315 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000156_160 |
0.2788097245 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 20 |
Hb_001021_160 |
0.2788875874 |
- |
- |
PREDICTED: homeobox protein LUMINIDEPENDENS [Jatropha curcas] |