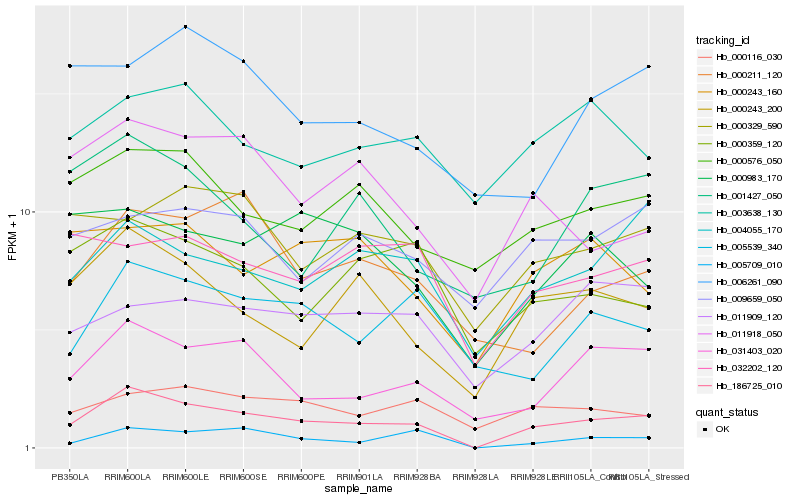
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186725_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_031403_020 |
0.161878561 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 3 |
Hb_000243_200 |
0.1657294714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005709_010 |
0.1660552384 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000576_050 |
0.1840721494 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 6 |
Hb_004055_170 |
0.1844193693 |
- |
- |
nucleosome assembly protein, putative [Ricinus communis] |
| 7 |
Hb_005539_340 |
0.1870775456 |
- |
- |
PREDICTED: CDT1-like protein a, chloroplastic [Jatropha curcas] |
| 8 |
Hb_011918_050 |
0.1872736465 |
- |
- |
- |
| 9 |
Hb_000329_590 |
0.1892487842 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000211_120 |
0.1895617898 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 11 |
Hb_000116_030 |
0.1897705571 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 12 |
Hb_000359_120 |
0.1942202217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001427_050 |
0.1950909623 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 14 |
Hb_000243_160 |
0.1956712805 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 15 |
Hb_000983_170 |
0.1959710606 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_032202_120 |
0.1967558189 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006261_090 |
0.1972548617 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 18 |
Hb_011909_120 |
0.1978390794 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 19 |
Hb_009659_050 |
0.198183092 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 20 |
Hb_003638_130 |
0.1987934051 |
- |
- |
PREDICTED: uncharacterized protein LOC105645779 [Jatropha curcas] |