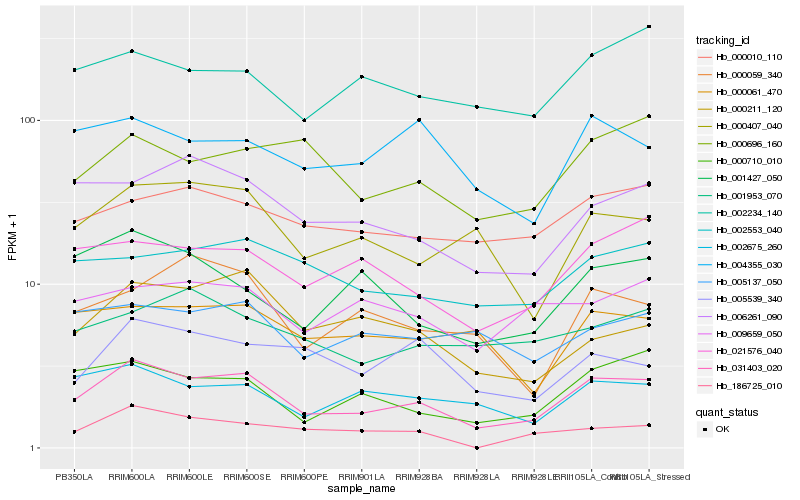
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031403_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 2 |
Hb_006261_090 |
0.1414425248 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 3 |
Hb_000710_010 |
0.1422469561 |
- |
- |
PREDICTED: uncharacterized protein LOC105630356 [Jatropha curcas] |
| 4 |
Hb_021576_040 |
0.1430995478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002675_260 |
0.1439160556 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 6 |
Hb_000407_040 |
0.1478751328 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002553_040 |
0.1502449179 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 8 |
Hb_000010_110 |
0.1505157663 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001427_050 |
0.1521445478 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 10 |
Hb_000211_120 |
0.1547641345 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 11 |
Hb_002234_140 |
0.1553828957 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 12 |
Hb_000696_160 |
0.1575259964 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 13 |
Hb_005137_050 |
0.1597714463 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 14 |
Hb_004355_030 |
0.1600943085 |
- |
- |
fumarylacetoacetate hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000059_340 |
0.1606285361 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 16 |
Hb_005539_340 |
0.1610241284 |
- |
- |
PREDICTED: CDT1-like protein a, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001953_070 |
0.1611459691 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 18 |
Hb_009659_050 |
0.1612514416 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 19 |
Hb_000061_470 |
0.1615774587 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 20 |
Hb_186725_010 |
0.161878561 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |