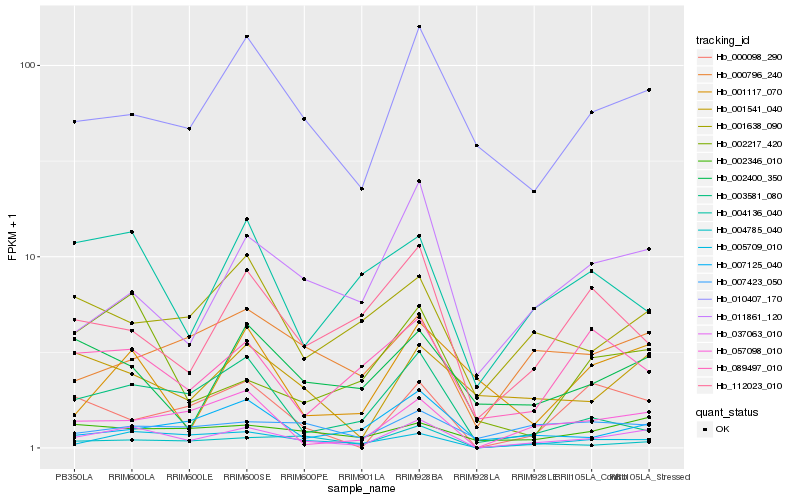
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_037063_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005709_010 |
0.1952383181 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_057098_010 |
0.2029253001 |
- |
- |
- |
| 4 |
Hb_010407_170 |
0.2144818354 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 5 |
Hb_004785_040 |
0.2201176131 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Jatropha curcas] |
| 6 |
Hb_011861_120 |
0.2215548291 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 7 |
Hb_001541_040 |
0.2297722117 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000796_240 |
0.2345789902 |
- |
- |
- |
| 9 |
Hb_002346_010 |
0.2348272233 |
- |
- |
- |
| 10 |
Hb_001638_090 |
0.2422331737 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 11 |
Hb_003581_080 |
0.2433646537 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 12 |
Hb_002400_350 |
0.2435153094 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_000098_290 |
0.2455614677 |
- |
- |
- |
| 14 |
Hb_089497_010 |
0.2461194299 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_004136_040 |
0.246286875 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 16 |
Hb_007125_040 |
0.2479449555 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_112023_010 |
0.2489320075 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 18 |
Hb_002217_420 |
0.24903371 |
- |
- |
hypothetical protein EUTSA_v10021493mg [Eutrema salsugineum] |
| 19 |
Hb_007423_050 |
0.2533077492 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 3 [Jatropha curcas] |
| 20 |
Hb_001117_070 |
0.2589333989 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |