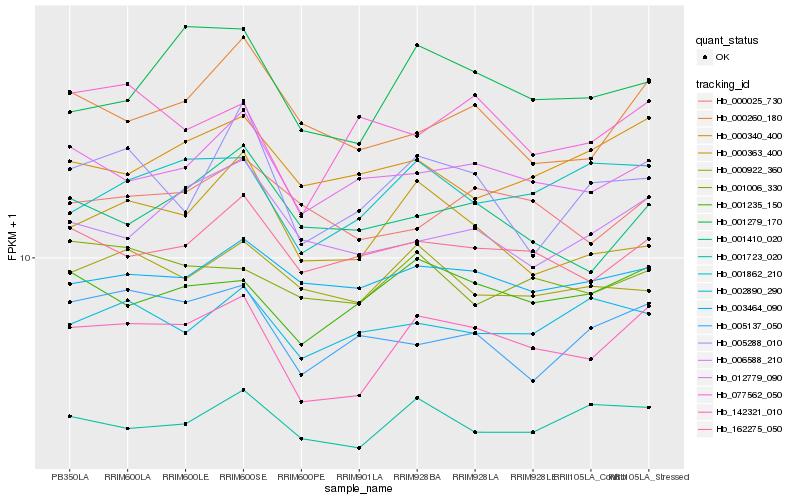
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142321_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 2 |
Hb_001235_150 |
0.0952775997 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012779_090 |
0.0981461732 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 4 |
Hb_001862_210 |
0.0984365979 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 5 |
Hb_001279_170 |
0.1010700201 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001723_020 |
0.1011467591 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 7 |
Hb_162275_050 |
0.1019567611 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 8 |
Hb_000363_400 |
0.1114590726 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 9 |
Hb_003464_090 |
0.1115554067 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 10 |
Hb_001006_330 |
0.1145872036 |
- |
- |
sodium/hydrogen exchanger plant, putative [Ricinus communis] |
| 11 |
Hb_006588_210 |
0.1146257651 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 12 |
Hb_000922_360 |
0.1147180014 |
- |
- |
- |
| 13 |
Hb_005137_050 |
0.114966794 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 14 |
Hb_000260_180 |
0.1152545377 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 15 |
Hb_001410_020 |
0.1154469441 |
- |
- |
cdk8, putative [Ricinus communis] |
| 16 |
Hb_077562_050 |
0.1162595039 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 17 |
Hb_000025_730 |
0.1163603604 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000340_400 |
0.1163958002 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 19 |
Hb_002890_290 |
0.1168708576 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005288_010 |
0.1173842352 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |