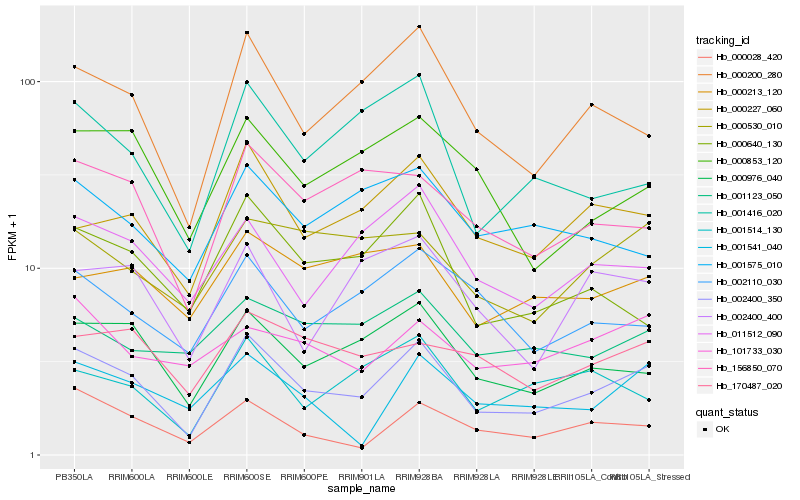
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_350 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 2 |
Hb_000200_280 |
0.1297100336 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 3 |
Hb_000976_040 |
0.131589596 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 4 |
Hb_000227_060 |
0.1483241564 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 5 |
Hb_156850_070 |
0.150791675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001416_020 |
0.1545887647 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 7 |
Hb_000028_420 |
0.155183051 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_011512_090 |
0.1561106798 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 9 |
Hb_000853_120 |
0.1588571393 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 10 |
Hb_000640_130 |
0.1611536485 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_170487_020 |
0.1623516055 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 12 |
Hb_000530_010 |
0.1629753235 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 13 |
Hb_002110_030 |
0.1636630414 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_002400_400 |
0.1666215423 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_001514_130 |
0.1670121232 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 16 |
Hb_001541_040 |
0.1694208738 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001575_010 |
0.1707701183 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_101733_030 |
0.1720854603 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 19 |
Hb_001123_050 |
0.1733872133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000213_120 |
0.1740997263 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |