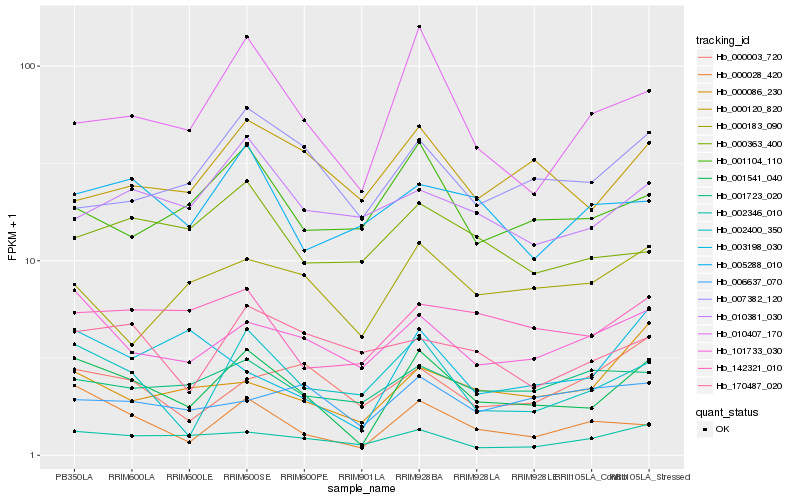
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_040 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000028_420 |
0.1495131124 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_101733_030 |
0.1642177028 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 4 |
Hb_010407_170 |
0.1645638617 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 5 |
Hb_002400_350 |
0.1694208738 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 6 |
Hb_002346_010 |
0.1703987354 |
- |
- |
- |
| 7 |
Hb_142321_010 |
0.1823342093 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 8 |
Hb_001104_110 |
0.1905798778 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 9 |
Hb_001723_020 |
0.192341529 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 10 |
Hb_000086_230 |
0.1929268542 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000183_090 |
0.1966287741 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000120_820 |
0.1986861 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 13 |
Hb_003198_030 |
0.1988202985 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_005288_010 |
0.1996024309 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_007382_120 |
0.2009042837 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000363_400 |
0.2010796898 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 17 |
Hb_000003_720 |
0.2011943214 |
- |
- |
hypothetical protein POPTR_0003s04720g [Populus trichocarpa] |
| 18 |
Hb_006637_070 |
0.2031067244 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 19 |
Hb_170487_020 |
0.2040489438 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 20 |
Hb_010381_030 |
0.2046572839 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |