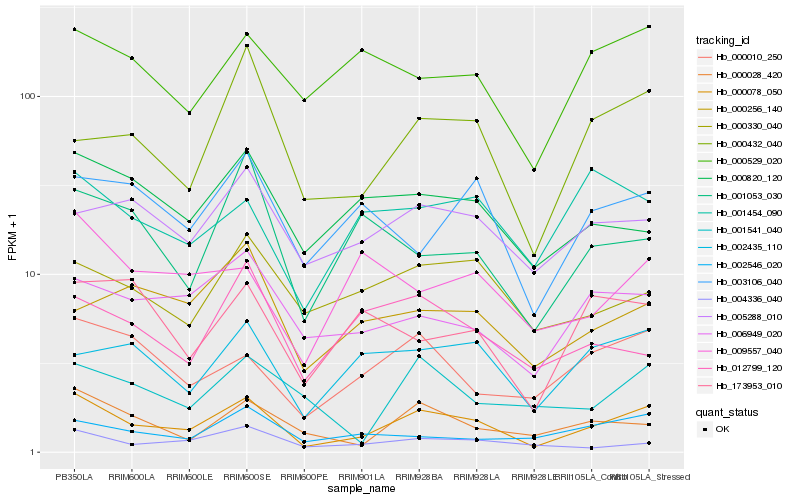
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000078_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 2 |
Hb_000010_250 |
0.1806523159 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 3 |
Hb_000432_040 |
0.1868739064 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 4 |
Hb_002435_110 |
0.1869749591 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 5 |
Hb_000028_420 |
0.1872280378 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_006949_020 |
0.1874976582 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine-C(5))-methyltransferase isoform X1 [Jatropha curcas] |
| 7 |
Hb_004336_040 |
0.1907407824 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 8 |
Hb_000820_120 |
0.1958398665 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 9 |
Hb_005288_010 |
0.1975539587 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000529_020 |
0.1987356157 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 11 |
Hb_000330_040 |
0.2002526082 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 12 |
Hb_002546_020 |
0.2018292165 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Malus domestica] |
| 13 |
Hb_003106_040 |
0.2040975441 |
- |
- |
lipase, putative [Ricinus communis] |
| 14 |
Hb_001454_090 |
0.2050496576 |
- |
- |
PREDICTED: alanyl-tRNA editing protein Aarsd1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001541_040 |
0.2071136404 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000256_140 |
0.2076590654 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009557_040 |
0.2089931097 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 18 |
Hb_001053_030 |
0.2090293397 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_173953_010 |
0.2095092182 |
- |
- |
hypothetical protein RCOM_1267370 [Ricinus communis] |
| 20 |
Hb_012799_120 |
0.2109392363 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |