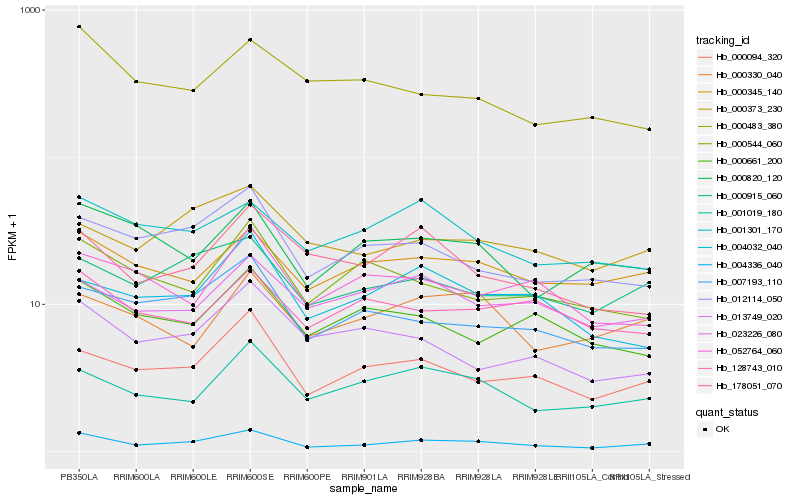
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004336_040 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 2 |
Hb_000094_320 |
0.1153385717 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 3 |
Hb_001019_180 |
0.124503481 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 4 |
Hb_000915_060 |
0.1287639841 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 5 |
Hb_000345_140 |
0.1311197231 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 6 |
Hb_012114_050 |
0.1317682854 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007193_110 |
0.1365372023 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 8 |
Hb_000820_120 |
0.1425182275 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 9 |
Hb_000373_230 |
0.1436187283 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000330_040 |
0.1438835049 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 11 |
Hb_004032_040 |
0.1443858775 |
- |
- |
unknown [Medicago truncatula] |
| 12 |
Hb_178051_070 |
0.1451901136 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 13 |
Hb_023226_080 |
0.1457281685 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000544_060 |
0.1466705118 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 15 |
Hb_128743_010 |
0.1483939463 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_013749_020 |
0.1488906321 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_001301_170 |
0.1498447957 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 18 |
Hb_000661_200 |
0.1513348542 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 19 |
Hb_052764_060 |
0.1524244879 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000483_380 |
0.1536971063 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |